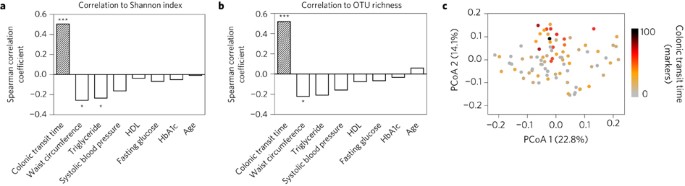
Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Little is known about how colonic transit time relates to human colonic metabolism and its importance for host health, although a firm stool consistency, a proxy for a long colonic
transit time, has recently been positively associated with gut microbial richness. Here, we show that colonic transit time in humans, assessed using radio-opaque markers, is associated with
overall gut microbial composition, diversity and metabolism. We find that a long colonic transit time associates with high microbial richness and is accompanied by a shift in colonic
metabolism from carbohydrate fermentation to protein catabolism as reflected by higher urinary levels of potentially deleterious protein-derived metabolites. Additionally, shorter colonic
transit time correlates with metabolites possibly reflecting increased renewal of the colonic mucosa. Together, this suggests that a high gut microbial richness does not _per se_ imply a
healthy gut microbial ecosystem and points at colonic transit time as a highly important factor to consider in microbiome and metabolomics studies. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 digital issues and online
access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local
taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING
VIEWED BY OTHERS GUT PHYSIOLOGY AND ENVIRONMENT EXPLAIN VARIATIONS IN HUMAN GUT MICROBIOME COMPOSITION AND METABOLISM Article Open access 27 November 2024 DELVING THE DEPTHS OF ‘TERRA
INCOGNITA’ IN THE HUMAN INTESTINE — THE SMALL INTESTINAL MICROBIOTA Article 23 October 2024 PROFILING THE HUMAN INTESTINAL ENVIRONMENT UNDER PHYSIOLOGICAL CONDITIONS Article Open access 10
May 2023 REFERENCES * Le Chatelier, E. _et al._ Richness of human gut microbiome correlates with metabolic markers. _Nature_ 500, 541–546 (2013). Article Google Scholar * Lozupone, C. A.,
Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. _Nature_ 489, 220–230 (2012). Article Google Scholar *
Vandeputte, D. _et al._ Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. _Gut_ 65, 57–62 (2015). Article Google
Scholar * Tigchelaar, E. F. _et al._ Gut microbiota composition associated with stool consistency. _Gut_ 65, 540–542 (2016). Article Google Scholar * Wang, Y. T. _et al._ Regional
gastrointestinal transit and pH studied in 21 healthy volunteers using the wireless motility capsule: influence of age, gender, study country and testing protocol. _Aliment. Pharmacol.
Ther._ 42, 761–772 (2015). Article Google Scholar * Nyangale, E. P., Mottram, D. S. & Gibson, G. R. Gut microbial activity, implications for health and disease: the potential role of
metabolite analysis. _J. Proteome Res._ 11, 5573–5585 (2012). Article Google Scholar * Tremaroli, V. & Bäckhed, F. Functional interactions between the gut microbiota and host
metabolism. _Nature_ 489, 242–249 (2012). Article Google Scholar * Davila, A.-M. _et al._ Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host.
_Pharmacol. Res._ 68, 95–107 (2013). Article Google Scholar * Russell, W. R., Hoyles, L., Flint, H. J. & Dumas, M.-E. Colonic bacterial metabolites and human health. _Curr. Opin.
Microbiol._ 16, 246–254 (2013). Article Google Scholar * Andriamihaja, M. _et al._ The deleterious metabolic and genotoxic effects of the bacterial metabolite p-cresol on colonic
epithelial cells. _Free Radic. Biol. Med._ 85, 219–227 (2015). Article Google Scholar * Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal
cancer. _Nature Rev. Microbiol._ 12, 661–672 (2014). Article Google Scholar * Shafi, T. _et al._ Free levels of selected organic solutes and cardiovascular morbidity and mortality in
hemodialysis patients: results from the retained organic solutes and clinical outcomes (ROSCO) investigators. _PLoS ONE_ 10, e0126048 (2015). Article Google Scholar * Gabriele, S. _et al._
Urinary p-cresol is elevated in young French children with autism spectrum disorder: a replication study. _Biomarkers_ 19, 463–470 (2014). Article Google Scholar * Ibrügger, S. _et al._
Two randomized cross-over trials assessing the impact of dietary gluten or wholegrain on the gut microbiome and host metabolic health. _J. Clin. Trials_ 4, 1000178 (2014). Article Google
Scholar * Falony, G. _et al._ Population-level analysis of gut microbiome variation. _Science_ 352, 560–564 (2016). Article Google Scholar * Arumugam, M. _et al._ Enterotypes of the human
gut microbiome. _Nature_ 473, 174–180 (2011). Article Google Scholar * Roager, H. M., Licht, T. R., Poulsen, S. K., Larsen, T. M. & Bahl, M. I. Microbial enterotypes, inferred by the
prevotella-to-bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. _Appl. Environ. Microbiol._ 80, 1142–1149 (2014). Article
Google Scholar * Claus, S. P. _et al._ Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. _Mol. Syst. Biol._ 4, 219 (2008). Article Google Scholar
* He, X. & Slupsky, C. M. Metabolic fingerprint of dimethyl sulfone (DMSO2) in microbial-mammalian co-metabolism. _J. Proteome Res._ 13, 5281–5292 (2014). Article Google Scholar *
Kilkkinen, A. _et al._ Use of oral antimicrobials decreases serum enterolactone concentration. _Am. J. Epidemiol._ 155, 472–477 (2002). Article Google Scholar * Topp, H., Sander, G.,
Heller-Schöch, G. & Schöch, G. Determination of 7-methylguanine, _N_2,_N_2-dimethylguanosine, and pseudouridine in ultrafiltrated serum of healthy adults by high-performance liquid
chromatography. _Anal. Biochem._ 161, 49–56 (1987). Article Google Scholar * Topp, H. & Schöch, G. Whole-body degradation rates of transfer-, ribosomal-, and messenger ribonucleic
acids and resting metabolic rate in 3- to 18-year-old humans. _Pediatr. Res._ 47, 163–163 (2000). Article Google Scholar * Mirvish, S. S., Medalie, J., Linsell, C. A., Yousuf, E. &
Reyad, S. 7-methylguanine and other minor urinary purines: values for normal subjects from Israel, Gaza, and Kenya, and for patients with cancer of various organs or cirrhosis of the liver.
_Cancer_ 27, 736–743 (1971). Article Google Scholar * Johansson, M. E. V., Larsson, J. M. H. & Hansson, G. C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the
outer layer is a legislator of host–microbial interactions. _Proc. Natl Acad. Sci. USA_ 108(Suppl), 4659–4665 (2011). Article CAS Google Scholar * Barrios, C. _et al._
Gut-microbiota-metabolite axis in early renal function decline. _PLoS ONE_ 10, e0134311 (2015). Article Google Scholar * Pimentel, M. _et al._ Methane, a gas produced by enteric bacteria,
slows intestinal transit and augments small intestinal contractile activity. _Am. J. Physiol. Gastrointest. Liver Physiol._ 290, G1089–G1095 (2006). Article Google Scholar * Soret, R. _et
al._ Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. _Gastroenterology_ 138, 1772–1782 (2010). Article Google Scholar * Degen, L. P.
& Phillips, S. F. How well does stool form reflect colonic transit? _Gut_ 39, 109–113 (1996). Article Google Scholar * Macfarlane, G. T., Cummings, J. H., Macfarlane, S. & Gibson,
G. R. Influence of retention time on degradation of pancreatic enzymes by human colonic bacteria grown in a 3-stage continuous culture system. _J. Appl. Bacteriol._ 67, 520–527 (1989).
Article Google Scholar * Macfarlane, S., Quigley, M., Hopkins, M., Newton, D. F. & Macfarlane, G. Polysaccharide degradation by human intestinal bacteria during growth under
multi-substrate limiting conditions in a three-stage continuous culture system. _FEMS Microbiol. Ecol._ 26, 231–243 (1998). Article Google Scholar * Cummings, J. H., Hill, M. J., Bone, E.
S., Branch, W. J. & Jenkins, D. J. The effect of meat protein and dietary fiber on colonic function and metabolism. II: Bacterial metabolites in feces and urine. _Am. J. Clin. Nutr._ 32,
2094–2101 (1979). Article Google Scholar * Benus, R. F. J. _et al._ Association between _Faecalibacterium prausnitzii_ and dietary fibre in colonic fermentation in healthy human subjects.
_Br. J. Nutr._ 104, 693–700 (2010). Article Google Scholar * Manach, C., Scalbert, A., Morand, C., Remesy, C. & Jimenez, L. Polyphenols: food sources and bioavailability. _Am. J.
Clin. Nutr._ 79, 727–747 (2004). Article Google Scholar * van Duynhoven, J. _et al._ Metabolic fate of polyphenols in the human superorganism. _Proc. Natl Acad. Sci. USA_ 108(Suppl),
4531–4538 (2011). Article CAS Google Scholar * Gross, G. _et al._ _In vitro_ bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays
strong interindividual variability. _J. Agric. Food Chem._ 58, 10236–10246 (2010). Article Google Scholar * Bode, L. M. _et al._ _In vivo_ and _in vitro_ metabolism of trans-resveratrol by
human gut microbiota. _Am. J. Clin. Nutr._ 97, 295–309 (2013). Article Google Scholar * Shimotoyodome, A., Meguro, S., Hase, T., Tokimitsu, I. & Sakata, T. Decreased colonic mucus in
rats with loperamide-induced constipation. _Comp. Biochem. Physiol. A. Mol. Integr. Physiol._ 126, 203–212 (2000). Article Google Scholar * Toden, S., Bird, A. R., Topping, D. L. &
Conlon, M. A. Resistant starch attenuates colonic DNA damage induced by higher dietary protein in rats. _Nutr. Cancer_ 51, 45–51 (2005). Article Google Scholar * Ten Bruggencate, S. J. M.,
Bovee-Oudenhoven, I. M. J., Lettink-Wissink, M. L. G., Katan, M. B. & Van Der Meer, R. Dietary fructo-oligosaccharides and inulin decrease resistance of rats to salmonella: protective
role of calcium. _Gut_ 53, 530–535 (2004). Article Google Scholar * Rao, J. N. & Wang, J.-Y. in _Molecule to Function to Disease_ (eds Granger, N., Granger, J. & Princeton, N. )
11–114 (Morgan & Claypool, 2011). Google Scholar * Sakata, T. Effects of indigestible dietary bulk and short chain fatty acids on the tissue weight and epithelial cell proliferation
rate of the digestive tract in rats. _J. Nutr. Sci. Vitaminol. (Tokyo)_ 32, 355–362 (1986). Article Google Scholar * Goodlad, R. A. _et al._ Effects of an elemental diet, inert bulk and
different types of dietary fibre on the response of the intestinal epithelium to refeeding in the rat and relationship to plasma gastrin, enteroglucagon, and PYY concentrations. _Gut_ 28,
171–180 (1987). Article Google Scholar * Lewis, S. J. & Heaton, K. W. Increasing butyrate concentration in the distal colon by accelerating intestinal transit. _Gut_ 41, 245–251
(1997). Article Google Scholar * Timmons, J., Chang, E. T., Wang, J.-Y. & Rao, J. N. Polyamines and gut mucosal homeostasis. _J. Gastrointest. Dig. Syst._ 2(Suppl 7), 001 (2012).
PubMed PubMed Central Google Scholar * Earle, K. A. _et al._ Quantitative imaging of gut microbiota spatial organization. _Cell Host Microbe_ 18, 478–488 (2015). Article Google Scholar
* Guérin, A. _et al._ Risk of developing colorectal cancer and benign colorectal neoplasm in patients with chronic constipation. _Aliment. Pharmacol. Ther._ 40, 83–92 (2014). Article Google
Scholar * Schadt, S., Chen, L.-Z. & Bischoff, D. Evaluation of relative LC/MS response of metabolites to parent drug in LC/nanospray ionization mass spectrometry: potential
implications in MIST assessment. _J. Mass Spectrom._ 46, 1281–1286 (2011). Article Google Scholar * McKeown, C., Hisle-Gorman, E., Eide, M., Gorman, G. H. & Nylund, C. M. Association
of constipation and fecal incontinence with attention-deficit/hyperactivity disorder. _Pediatrics_ 132, e1210-5 (2013). Article Google Scholar * Pang, K. H. & Croaker, G. D. H.
Constipation in children with autism and autistic spectrum disorder. _Pediatr. Surg. Int._ 27, 353–358 (2011). Article Google Scholar * Wu, M.-J. _et al._ Colonic transit time in long-term
dialysis patients. _Am. J. Kidney Dis._ 44, 322–327 (2004). Article Google Scholar * Waller, P. A. _et al._ Dose–response effect of _Bifidobacterium lactis_ HN019 on whole gut transit
time and functional gastrointestinal symptoms in adults. _Scand. J. Gastroenterol._ 46, 1057–1064 (2011). Article Google Scholar * Abrahamsson, H. & Antov, S. Accuracy in assessment of
colonic transit time with particles: how many markers should be used? _Neurogastroenterol. Motil._ 22, 1164–1169 (2010). Article Google Scholar * Wesolowska-Andersen, A. _et al._ Choice
of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. _Microbiome_ 2, 19 (2014). Article Google Scholar * Godon, J.
J., Zumstein, E., Dabert, P., Habouzit, F. & Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. _Appl. Environ.
Microbiol._ 63, 2802–2813 (1997). Google Scholar * Klindworth, A. _et al._ Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based
diversity studies. _Nucleic Acids Res._ 41, e1 (2013). Article Google Scholar * Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. _Nature Methods_ 10,
996–998 (2013). Article Google Scholar * Haas, B. J. _et al._ Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. _Genome Res._ 21, 494–504
(2011). Article Google Scholar * Caporaso, J. G. _et al._ QIIME allows analysis of high-throughput community sequencing data. _Nature Methods_ 7, 335–336 (2010). Article Google Scholar *
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. _Bioinformatics_ 26, 2460–2461 (2010). Article Google Scholar * McDonald, D. _et al._ An improved Greengenes
taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. _ISME J._ 6, 610–618 (2012). Article Google Scholar * Caporaso, J. G. _et al._ PyNAST: a
flexible tool for aligning sequences to a template alignment. _Bioinformatics_ 26, 266–267 (2010). Article Google Scholar * Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree:
computing large minimum evolution trees with profiles instead of a distance matrix. _Mol. Biol. Evol._ 26, 1641–1650 (2009). Article Google Scholar * Chen, Y. _et al._ Combination of
injection volume calibration by creatinine and MS signals’ normalization to overcome urine variability in LC-MS-based metabolomics studies. _Anal. Chem._ 85, 7659–7665 (2013). Article
Google Scholar * Want, E. J. _et al._ Global metabolic profiling procedures for urine using UPLC-MS. _Nature Protoc._ 5, 1005–1018 (2010). Article Google Scholar * Smith, C. A., Want, E.
J., O'Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. _Anal.
Chem._ 78, 779–787 (2006). Article Google Scholar * Wishart, D. S. _et al._ HMDB 3.0—The human metabolome database in 2013. _Nucleic Acids Res._ 41, D801–D807 (2013). Article Google
Scholar * Kim, S. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. _Commun. Stat. Appl. Methods_ 22, 665–674 (2015). Google Scholar * Benjamini, Y.
& Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. _J. R. Stat. Soc. Ser. B_ 57, 289–300 (1995). Google Scholar * Warnes, G. R.
_et al._ gplots: various R programming tools for plotting data. R package version 2.16.0 (CRAN, 2015); http://cran.r-project.org/package=gplots * Oksanen, J. _et al._ vegan: community
ecology package. R package version 2.3.1 (CRAN, 2015); https://cran.r-project.org/package=vegan * Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R
language. _Bioinformatics_ 20, 289–290 (2004). Article Google Scholar Download references ACKNOWLEDGEMENTS The authors thank K.V. Vibefelt for helping out with DNA extraction and N. Bicen
for performing the PCR and sequencing. The sequencing was carried out by the DTU in-house facility (DTU Multi-Assay Core, DMAC), Technical University of Denmark. This work was funded by the
Danish Council for Strategic Research (grant no. 11-116163; Center for Gut, Grain and Greens), by the Technical University of Denmark and by the personal Danisco Award (to T.R.L.). The Novo
Nordisk Foundation Center for Basic Metabolic Research is an independent research centre at the University of Copenhagen and is partly funded by an unrestricted donation from the Novo
Nordisk Foundation. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * National Food Institute, Technical University of Denmark, DK-2860 Søborg, Denmark Henrik M. Roager, Martin I. Bahl, Henrik
L. Frandsen, Vera Carvalho & Tine R. Licht * Department of Systems Biology, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark Lea B. S. Hansen, Marlene D. Dalgaard, Damian R.
Plichta, Thomas Sicheritz-Pontén, H. Bjørn Nielsen & Ramneek Gupta * The Novo Nordisk Foundation Center for Basic Metabolic Research, Section of Metabolic Genetics, University of
Copenhagen, DK-2200 København N, Denmark Rikke J. Gøbel, Henrik Vestergaard, Torben Hansen & Oluf Pedersen * Department of Radiology, Bispebjerg Hospital, DK-2400 København NV, Denmark
Morten H. Sparholt * Department of Nutrition, Exercise and Sport, University of Copenhagen, DK-1958 Frederiksberg C, Denmark Lotte Lauritzen & Mette Kristensen Authors * Henrik M. Roager
View author publications You can also search for this author inPubMed Google Scholar * Lea B. S. Hansen View author publications You can also search for this author inPubMed Google Scholar
* Martin I. Bahl View author publications You can also search for this author inPubMed Google Scholar * Henrik L. Frandsen View author publications You can also search for this author
inPubMed Google Scholar * Vera Carvalho View author publications You can also search for this author inPubMed Google Scholar * Rikke J. Gøbel View author publications You can also search for
this author inPubMed Google Scholar * Marlene D. Dalgaard View author publications You can also search for this author inPubMed Google Scholar * Damian R. Plichta View author publications
You can also search for this author inPubMed Google Scholar * Morten H. Sparholt View author publications You can also search for this author inPubMed Google Scholar * Henrik Vestergaard
View author publications You can also search for this author inPubMed Google Scholar * Torben Hansen View author publications You can also search for this author inPubMed Google Scholar *
Thomas Sicheritz-Pontén View author publications You can also search for this author inPubMed Google Scholar * H. Bjørn Nielsen View author publications You can also search for this author
inPubMed Google Scholar * Oluf Pedersen View author publications You can also search for this author inPubMed Google Scholar * Lotte Lauritzen View author publications You can also search
for this author inPubMed Google Scholar * Mette Kristensen View author publications You can also search for this author inPubMed Google Scholar * Ramneek Gupta View author publications You
can also search for this author inPubMed Google Scholar * Tine R. Licht View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.I.B., R.J.G.,
H.V., T.H., T.S.-P., O.P., L.L., M.K., R.G. and T.R.L. assembled the cohort and developed protocols and infrastructure to obtain the biological samples and clinical metadata. M.H.S. measured
the colonic transit time. L.B.S.H., V.C. and M.D.D. prepared the faecal samples, extracted DNA and performed 16S rRNA gene sequencing. L.B.S.H. performed the 16S data analyses, with
contributions from V.C. H.M.R. and H.L.F. prepared the urine samples, performed the urine metabolic profiling and identified the urinary metabolites. H.M.R. performed the statistical
correlation analyses with contributions from L.B.S.H., D.R.P. and H.B.N. H.M.R., M.I.B., H.B.N., L.B.S.H., T.S.-P., R.G., M.K. and T.R.L. interpreted the data. H.M.R. and T.R.L. wrote the
manuscript and all authors read, revised and approved the final manuscript. CORRESPONDING AUTHOR Correspondence to Tine R. Licht. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare
no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Tables 1-6, Supplementary Figures 1-14 and Supplementary References. (PDF 2035 kb) RIGHTS
AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Roager, H., Hansen, L., Bahl, M. _et al._ Colonic transit time is related to bacterial metabolism and mucosal
turnover in the gut. _Nat Microbiol_ 1, 16093 (2016). https://doi.org/10.1038/nmicrobiol.2016.93 Download citation * Received: 15 December 2015 * Accepted: 20 May 2016 * Published: 27 June
2016 * DOI: https://doi.org/10.1038/nmicrobiol.2016.93 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
