Sox9 expression decreases survival of patients with intrahepatic cholangiocarcinoma by conferring chemoresistance
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT BACKGROUND Sex-determining region Y-box (SRY-box) containing gene 9 (SOX9) expression confers cancer stem cell features. However, SOX9 function in intrahepatic cholangiocarcinoma
(iCCA) is unknown. This study investigated the effects and underlying mechanisms of SOX9 in iCCA. METHODS SOX9 expression in 59 iCCA patients was examined by immunohistochemistry. The
association between SOX9 expression and clinical outcome was evaluated. Gene signature and biological functions of SOX9 in iCCA were examined in vitro. RESULTS iCCA patients with high SOX9
expression had shorter survival time than those with low SOX9. In patients receiving chemotherapy, median survival time in patients with low and high levels of SOX9 were 62 and 22 months,
respectively. In vitro, gemcitabine increased SOX9 expression in iCCA cells. When SOX9 was knocked down, gemcitabine-induced apoptosis was markedly increased. Silencing SOX9 significantly
inhibited gemcitabine-induced phosphorylation of checkpoint kinase 1, a key cell cycle checkpoint protein that coordinates the DNA damage response and inhibited the expression of multidrug
resistance genes. Microarray analyses showed that SOX9 knockdown in CCA cells altered gene signatures associated with multidrug resistance and p53 signalling. CONCLUSIONS SOX9 governs the
response of CCA cells to chemotherapy. SOX9 is a biomarker to select iCCA patients eligible for efficient chemotherapy. SIMILAR CONTENT BEING VIEWED BY OTHERS WNT-TCF7-SOX9 AXIS PROMOTES
CHOLANGIOCARCINOMA PROLIFERATION AND PEMIGATINIB RESISTANCE IN A FGF7-FGFR2 AUTOCRINE PATHWAY Article 15 April 2022 THE HETEROGENEITY OF SIGNALING PATHWAYS AND DRUG RESPONSES IN INTRAHEPATIC
CHOLANGIOCARCINOMA WITH DISTINCT GENETIC MUTATIONS Article Open access 11 January 2024 SINGLE-CELL TRANSCRIPTOMIC ANALYSIS SUGGESTS TWO MOLECULARLY DISTINCT SUBTYPES OF INTRAHEPATIC
CHOLANGIOCARCINOMA Article Open access 28 March 2022 INTRODUCTION Cholangiocarcinoma (CCA) is the second most common primary liver cancer following hepatocellular carcinoma (HCC), and
accounts for approximately 10–15% of all primary liver malignancies.1 The global incidence and mortality rate for CCA have been increasing over the past decades.2,3 Anatomically, CCA is
classified into intrahepatic (iCCA) and extrahepatic cholangiocarcinoma (eCCA) depending on the location of the tumour along the biliary tract.4 To date, curative surgical resection is the
most efficient treatment for long-term survival of selected iCCA patients.5,6 However, in most cases, the tumours are quite advanced at the time of diagnosis and surgical resection is not
possible.5 Systemic chemotherapy and radiotherapy regimens remain the only approach to render patient eligible for surgery and palliative treatment.7 However, the response of iCCA to these
treatments is very weak.7 Therefore, elucidating the underlying mechanisms of iCCA chemoresistance is one key issue to improve survival of patients. Sex-determining region Y-box (SRY-box)
containing gene 9 (SOX9) belongs to the SOX family of transcription factors.8 It is widely expressed in multiple organs during embryonic development, including the liver.9,10 In liver
embryogenesis, SOX9 expression is the most specific and earliest marker of hepatoblasts and determines the timing of intrahepatic bile duct morphogenesis.11,12 In normal adult liver, SOX9 is
expressed in the periportal small intrahepatic ducts, and peribiliary glands lining the large bile ducts.13 SOX9 plays important roles in maintaining liver homeostasis, regulating liver
regeneration, and eventually in liver cancer development.14 In acute or chronic liver disease, SOX9 expression robustly manifests in ductular reactions (DRs), which contain putative
progenitor cells capable of differentiating into both cholangiocytes and hepatocytes.15 Moreover, SOX9-positive cells express stem cell markers, such as epithelial cell adhesion molecule
(EpCAM), neural cell adhesion molecule, CD133 and CXC motif chemokine receptor 4.13,16 In contrast to normal hepatocytes, where SOX9 is not expressed, a subset of HCC cells displayed SOX9
expression. These patients usually demonstrate severe venous cancer invasion, advanced tumour stage and shorter survival.17 Recent studies reported that SOX9-positive HCC cells exhibit liver
cancer stem cell (CSC)-like features, and that SOX9 in cancer cells confers self-renewal and tumourigenicity by promoting symmetrical cell division.17,18 To date, only few studies had
addressed the role of SOX9 in CCA.19 Here, we report clinical and functional data supporting an oncogenic role and therapeutic significance of SOX9 expression in iCCA. MATERIALS AND METHODS
PATIENTS AND LIVER TISSUES This study enroled 59 iCCA patients from Tübingen, Germany (18 iCCA patients) and Rennes, France (41 iCCA patients) between 2002 and 2010. In addition, 21 liver
tissues from patients with chronic hepatitis B infection were enroled in Mannheim, Germany. Basic characteristics of the enroled chronic hepatitis B and iCCA patients are shown in
Supplementary Tables 1 and 2. The study protocol fulfilled national laws and regulations and was approved by the local Ethics Committees. CELL CULTURE AND TREATMENT The following cell lines
were investigated in the study: CC-SW-1 and HuCCT-1 (iCCA lines), EGI-1 and TFK-1 (eCCA lines), HCCC-9810 (mix CCA line) and MMNK-1 (normal cholangiocyte line). EGI-1, CC-SW-1 and MMNK-1
were cultured in Dulbecco's modified Eagle's medium (DMEM) (BE12-709F, Lonza) supplemented with 10% foetal bovine serum (FBS) (10270-098, Invitrogen), 4mM l-glutamine (17-605C,
Lonza) and 100 U/mL penicillin/streptomycin (A2210 Biochrom KG). TFK-1 and HuCCT-1 were cultured in RPMI-1640 supplemented with 10% FBS (10270-098, Invitrogen), 4 mM l-glutamine (17-605 C,
Lonza) and 100 U/mL penicillin/streptomycin (A2210 Biochrom KG). All the cell lines were cultured in a humidified incubator at 37 °C and with 5% CO2 atmosphere. Cells underwent starvation
without FBS medium for 10 to 16 h before treatment with gemcitabine and cisplatin (kindly provided by Prof. Lu LG, Shanghai Jiao Tong University School of Medicine), which was dissolved in
phosphate-buffered saline (PBS) to make a 100 mM stock solution and diluted with cell culture medium to indicated concentrations during treatment. IMMUNOHISTOCHEMISTRY AND STAINING
EVALUATION Tissue microarray assay was performed as previously described.20 In brief, formalin-fixed, paraffin-embedded specimens were deparaffinised in serial ethanol dilutions and
rehydrated. After a single PBS wash, heat-induced antigen retrieval was performed with 1 mM EDTA solution, pH 8.4 (03677; Sigma-Aldrich, Steinheim, Germany) at 98 °C for 10 min. Endogenous
peroxidase activity was blocked with Dako dual endogenous enzyme blocking reagent (S2003; Dako, Via Real, Carpinteria, CA, USA), followed by blocking with 3% hydrogen peroxidase for 5 min at
room temperature to prevent unspecific binding of antibodies. The tissue sections were incubated with polyclonal rabbit anti-SOX9 antibody (HPA001758; Sigma-Aldrich) at a dilution of 1:100,
or monoclonal mouse anti-cytokeratin 19 (CK19) antibody (SC-6287; Santa Cruz) at a dilution of 1:100 overnight at 4 °C. The specimens were subsequently washed in PBS for 3×5 min and
incubated with anti-rabbit or anti-mouse secondary antibody conjugated with horse radish peroxidase (HRP) for 1 h at room temperature, and then detected with 3,3′-diaminobenzidine for 7 min.
The slides were counterstained with haematoxylin. All sections were dehydrated and mounted with malinol mounting medium. Immunostaining results for SOX9 were scored semi-quantitatively
based on the intensity and proportion of positive tumour cell nuclei. In detail, the intensity score of SOX9 nuclear staining was defined as four grades: 0, negative; 1, weak with colour
yellow; 2, medium with colour brown; 3, strong with colour black. The number of cells with SOX9-positive nuclei was defined as six grades: 0, no detectable positive cells; 1, positive cells
≤1%; 2, positive cells >1%, and ≤10%; 3, positive cells >10%, and ≤33%; 4, positive cells > 33%, and ≤66%; 5, positive cells >66%. The final immune staining scores were
calculated as the intensity scores × the proportion scores. The samples with final scores over 10 were defined as “high SOX9 expression”, and the remainder as “low SOX9 expression”. The
representative pictures of SOX9 staining and for semi-quantitative scoring system are presented in Supplementary Figure 1. CK19 expression was categorised into high expression and low
expression according to the immunoreactivity in tumour cells. The immunoreactivity of CK19 was defined as four grades: 0, positive cells ≤1%; 1, positive cells >1% and ≤33%; 2, positive
cells >33% and ≤66%; 3, positive cells >66%. The samples with grade 3 were defined as high CK19 expression, and the others were low CK19 expression. RNA INTERFERENCE For transient
transfection of short interfering RNA (siRNA), cells were treated with indicated culture medium without penicillin/streptomycin. siRNA targeting human SOX9 (M-021507-00) and control siRNA
(D-001206-14) were purchased from Dharmacon. SOX9 siRNA were transfected with RNAiMAX (13778, Invitrogen). The transfection was performed in six-well cell culture vessels. Tumour cells were
plated at a density of 1.5 × 105 cells per well with 2 mL corresponding growth medium. Briefly, for siRNA transfection, 2 μl RNAiMAX was mixed with 20 pmol SOX9 siRNA in 200 μl Opti-MEM
medium. The mixtures were preincubated for 20 min at room temperature before adding to cells. RNA and whole-cell proteins were extracted 48 and 60 h after transfection for further
examination, respectively. MTT ASSAY Cells were incubated with 5 mg/mL 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) reagent (M5655, Sigma-Aldrich) for 5 h. Then, the
supernatant was removed carefully and the 100 µL solvent solution containing 40 µL of 10% sodium dodecyl sulphate (SDS), 40 µL dimethyl sulfoxide and 20 µL of 1.2% acetate acid solution (600
µL acetate acid in 50 mL PBS) was added and incubated overnight for measurement. Absorbance was measured at 570 nm with a reference to 630 nm. For cell viability assay and gemcitabine
half-maximal inhibitory concentration (IC50) measurement, cells were incubated in 96-well plate for 48 h before incubation with MTT. CELL CYCLE ANALYSIS Cells were harvested at 48 h after
siRNA treatment and washed with cold PBS, and then fixed with 70% cold ethanol. To remove RNA, the cells were re-suspended in solution containing Triton X-100 (0.1%) and 100 µg/mL RNase. The
samples were stained with propidium iodide (20 μg/mL) for 30 min in the dark, and then subjected to analysis for DNA content using FACS Calibur (BD Biosciences, Heidelberg, Germany) and
data analysis was performed using FlowJo version10 software. TRANSWELL MIGRATION ASSAY Cell culture inserts with 8 μM pore size (Falcon) were used. For tumour cell migration, 2.0 × 105 iCCA
tumour cells were suspended in RPMI or DMEM medium with 0.5% FBS and plated in the upper chambers. The lower chambers were filled RPMI or DMEM with 10% FBS. After 16 h, the medium in the
inserts were removed and washed with PBS. The inserts were filled with 3.7% formaldehyde for 5 min. Thereafter, the inserts were incubated in methanol for 30 min. The filters were stained
with 10% Giemsa (Sigma, St. Louis, MO, USA) for 15 min. The inner side was wiped with cotton swabs. Migrated cells were counted under a light microscope. CASPASE-3 ASSAY Caspase-3 assay was
performed as previously described.21 In brief, cells were lysed in 80 µL of lysis buffer (50 mM HEPES, 100 mM NaCl, 0.1% CHAPS, 1 mM DTT, 0.1 mM EDTA, pH 7.4). Then, 20 μl of cell lysate
were incubated in 70 μl reaction buffer (50 mM HEPES, 100 mM NaCl, 0.1% CHAPS, 10 mM DTT, 0. mM EDTA, 10% (w/v) glycerol, pH 7.4) and 10 μl AC-DEVD-AFC caspase-3 fluorimetric substrate
(Biomol, Hamburg, Germany) for 90 min at 37 °C. Subsequently, caspase-3 activity was detected by fluorometric measurement using Tecan infinite M200 (excitation 400 nm; emission 505 nm). The
caspase-3 activity was normalised to protein levels and reported as relative fluorescent units per minute per mg protein. IMMUNOBLOTTING Immunoblotting assay was performed as previously
described.20 Briefly, total cell protein was extracted on ice using radio immunoprecipitation assay buffer with freshly added protease and phosphatase inhibitors. Protein concentrations were
assessed with a Bio-Rad protein assay. Twenty micrograms of total cell protein extracts was subjected to 10% or 12% SDS-polyacrylamide electrophoresis gel and transferred to nitrocellulose
membranes. Five per cent non-fat milk in Tris-buffered saline with Tween-20 (TBST) was used to block nonspecific binding. Membranes were probed with primary and secondary antibodies in TBST
according to the manufacturer’s instructions. HRP-linked anti-mouse and anti-rabbit Abs were used as secondary antibodies. α-Tubulin and glyceraldehyde 3-phosphate dehydrogenase were used as
a loading control. Signal was visualised by incubating the blots in Supersignal Ultra (Pierce, Hamburg, Germany). RNA ISOLATION AND QUANTITATIVE REAL-TIME REVERSE TRANSCRIPTION POLYMERASE
CHAIN REACTION Total cell RNA was extracted using the InviTrap Spin Universal RNA Mini Kit (Stratec, Berlin, Germany), according to the manufacturer’s instructions. For first-strand
complementary DNA (cDNA) synthesis, reverse transcription of 500 ng RNA was performed with random primers (Thermo Scientific) and RevertAid H Minus M-MuLV reverse transcriptase (Thermo
Scientific) according to the manufacturer’s instructions and subsequently diluted with nuclease-free water (Invitrogen) to 10 ng/µL cDNA. For PCR amplification, 10.4 µL mixtures contained 5
µL (50 ng) template cDNA, 5 µL SYBR Green (4367659, Life Technologies) and 4 µM forward and reverse primer PCRs were run in triplicate and performed on a StepOnePlus Real-time PCR (Applied
Biosystems). PCR amplification cycling conditions comprised 10 min polymerase activation at 95 °C and 40 cycles at 95 °C for 15 s and 60 °C for 1 min. A melting-curve analysis was performed
for each PCR analysis. Relative quantification of target genes was normalised against the housekeeping gene _PPIA_. MICROARRAY Gene expression profiling was performed using arrays of human
HuGene-2_0-st-type from Affymetrix. Biotinylated antisense cDNA was prepared according to the Affymetrix standard labelling protocol with the GeneChip® WT Plus Reagent Kit and the GeneChip®
Hybridization, Wash and Stain Kit (both from Affymetrix, Santa Clara, CA, USA). Subsequently, the hybridisation on the chip was performed on a GeneChip Hybridization oven 640, then dyed in
the GeneChip Fluidics Station 450 and thereafter scanned with a GeneChip Scanner 3000. All of the equipment used was from the Affymetrix-Company (Affymetrix, High Wycombe, UK). BIOINFORMATIC
ANALYSES A Custom CDF Version 21 with ENTREZ-based gene definitions was used to annotate the arrays.22 The Raw fluorescence intensity values were normalised applying quantile normalisation
and RMA background correction. Differential expressed genes were identified by using a commercial software package SAS JMP10 Genomics, version 6, from SAS (SAS Institute, Cary, NC, USA). A
false-positive rate of _a_ = 0.05 with false discovery rate correction was taken as the level of significance. STATISTICAL ANALYSES Variables were summarised as means ± standard deviation
(SD) and depicted graphically as means ± SD. _P_ values were calculated using the _χ_2 test or calculated using a two-sided (unpaired) Student’s _t_ test. Kaplan–Meier survival curve and
univariate Cox analysis was used to evaluate overall survival (OS) rates and disease-free survival (DFS) rate of iCCA patients. _P_ values were calculated using the log-rank test. _P_ <
0.05 was considered significant. RESULTS SOX9 HAS DISTINCT EXPRESSION PATTERNS IN CHRONIC LIVER DISEASE AND CCA First, we compared expression of SOX9 and CK19, two classic markers of biliary
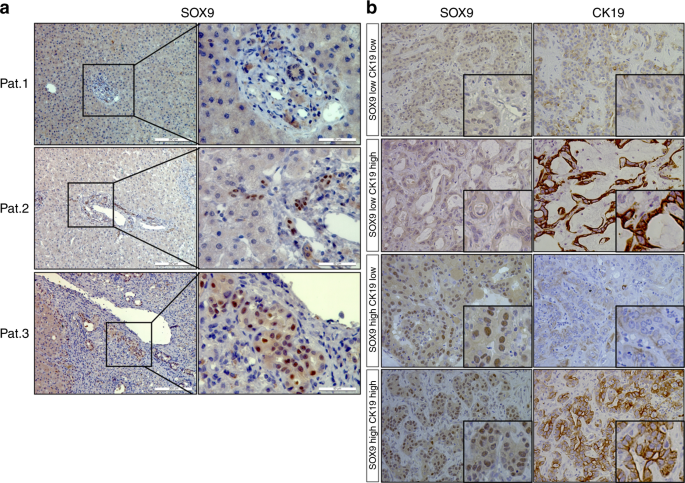
tree, in 80 patients with chronic liver disease or iCCA. Among 21 patients with chronic liver disease, 17 showed SOX9-positive immunoreactivity, whereas 4 were negative (Fig. 1a shows
representative patients). In contrast to SOX9, CK19 immunostaining was positive in all patients (data not shown). The results suggest that SOX9 expression in cholangiocytes is unstable in
chronic liver disease compared to CK19. Distinct from CK19, which localised in the cytoplasm of cholangiocytes, SOX9 was expressed in the nuclei of cells in the canals of Hering, reactive
ductules and bile ducts (Patients 1 and 2, Fig. 1a). As in chronic liver disease, SOX9 expression was observed in the nuclei of iCCA tumour cells, while CK19 localised in the cytoplasm of
cancer cells (Fig. 1b). Expressions of SOX9 and CK19 in cancer cells were heterogeneous. Figure 1b display four patterns of SOX9 and CK19 expression in iCCA: SOX9highCK19high,
SOX9highCK19low, SOX9lowCK19high and SOX9lowCK19low. There was no significant correlation between expression of SOX9 and CK19 in iCCA tumour cells (_P_ > 0.05). In all examined tissue
specimens, neither SOX9 nor CK19 were detected in hepatocytes. EXPRESSION OF SOX9 PREDICTS POOR CLINICAL OUTCOME OF ICCA Next, we analysed the correlation of SOX9 and CK19 expression with
clinical parameters of the iCCA patients, including age, gender, vascular invasion, existence of cirrhosis and American Joint Committee on Cancer (AJCC) classification. Among these clinical
parameters, CK19 expression was associated with AJCC classification of CCA, while SOX9 did not show any association with these clinical parameters (Supplementary Table 3). However,
multivariate analysis showed that among the analysed variables, only SOX9 expression significantly influenced the over survival (OS) of iCCA patients (hazard ratio = 3.614, 95% confidence
Interval = 1.493–9.076, _P_ = 0.006, Supplementary Table 4). Furthermore, Kaplan–Meier analysis and the log-rank test showed that patients with high SOX9 expression had shorter OS and
disease free survival (DFS) rates than those with low SOX9 expression (_P_ < 0.01 and _P_ < 0.05, respectively, Fig. 2a, c). The median OS time in patients with SOX9 low expression was
62 months, whereas the value in those patients with high SOX9 expression was only 22 months (Fig. 2a). In contrast to SOX9, there was no association between CK19 expression and survival
time in these patients (_P_ > 0.05, Fig. 2b, d). This cohort of iCCA included 9 patients who received chemotherapy (e.g. gemcitabine and cisplatin) (Supplementary Table 5). Among them,
six patients had low SOX9 expression and three patients showed high levels of SOX9. Survival analyses revealed that patients with high SOX9 expression had shorter OS time (_P_ < 0.05,
Fig. 3a). In patients who received chemotherapy, the mean survival time in patients with SOX9 low expression was 62 months, whereas the value in those patients with high SOX9 expression was
only 22 months (Fig. 3a). Except one patient who received chemotherapy following surgery, additional eight patients, five with low and three with high SOX9 expression, received chemotherapy
due to the recurrence of iCCA. The survival times until the end of the follow-up in five patients with low SOX9 levels were 16, 19, 20, 29 and 34 months, whereas the values in 3 with high
SOX9 expression were 13, 14 and 16 months, respectively (Supplementary Table 5). Survival difference between the two groups was significant (_P_ < 0.01, Fig. 3a). Notably, four out of
five patients with low SOX9 were still surviving when the follow-up ended (Supplementary Table 5). However, all three patients with high SOX9 expression were dead during follow-up
(Supplementary Table 5). CK19 expression did not show any correlation with chemotherapy response (_P_ > 0.05, data not shown). SOX9 INHIBITION SENSITISES CCA CELLS TO GEMCITABINE To
investigate how SOX9 expression in CCA cell might modify their response to chemotherapy, we performed microarray analysis in iCCA CC-SW-1 cells after SOX9 silencing. We found that gene
expression associated with drug metabolism and ABC transporters such as _ABCB1_ (MDR1) and _ABCC4_ (MRP4) was decreased, while genes related with the p53 signalling pathway were increased
when SOX9 was knocked down with siRNA (Supplementary Figure 2A). Western blot and quantitative polymerase chain reaction (qPCR) further confirmed that the expression of multidrug resistance
genes _ABCC4_ and _ABCB1_ was markedly reduced when SOX9 was inhibited in CC-SW-1 cells (Supplementary Figure 2B–C). Next, we treated different types of CCA cells with gemcitabine, an
analogue of deoxycytidine, which is widely used in the treatment of CCA. Notably, basal expression of SOX9 in CCA cells was significantly higher than in normal cholangiocytes (Fig. 4a). More
impressively, expression of SOX9 protein was further increased upon gemcitabine treatment in both iCCA CC-SW-1 and eCCA EGI-1 cells (Fig. 4b). To examine the function of SOX9 in
gemcitabine-treated CCA cells, we knocked down SOX9 expression using siRNA in CC-SW-1 and EGI-1 cells, followed by treatment with gemcitabine for 24 h (Fig. 4c). MTT assay showed that when
SOX9 expression was inhibited, the IC50 of gemcitabine-treated cells significantly decreased from 7.1 ± 0.15 to 2.0 ± 0.23 nM in CC-SW-1 cells and from 380.3 ± 249.1 to 46.3 ± 21.9 nM in
EGI-1 cells, respectively (Fig. 4d). Given the key role of phosphorylation of checkpoint kinase 1 (CHEK1) in coordinating the DNA damage response and inhibiting the expression of multidrug
resistance genes,23 we examined whether disruption of SOX9 impacted CHEK1 activation. Immunoblot analysis showed that SOX9 siRNA remarkably inhibited gemcitabine-dependent pCHEK1 in both
CC-SW-1 and EGI-1 cells and MRP4 expression in CC-SW-1 cells (Fig. 4d–f). Consistent with reduced expression of MRP4 and pCHK1, analyses based on immunoblot and cleaved caspase-3 activity
assay revealed marked increases in cleaved caspase-3 and caspase-8 expression and caspase activity in CCA cells with SOX9 knockdown, indicating that gemcitabine-induced apoptosis was
increased when SOX9 expression was inhibited (Fig. 4g, h). In addition to gemcitabine, we examined the role of SOX9 in cisplatin-treated CC-SW-1 and EGI cells. MTT assay showed that
knockdown of SOX9 did not have impact on cisplatin-inhibited cell viability in both cells (Supplementary Figure 3A–B). In contrast to gemcitabine, administration of cisplatin and/or
knockdown of SOX9 did not influence the expression of pCHEK1 (Supplementary Figure 3C–D). SOX9 IS ESSENTIAL FOR CCA CELL PROLIFERATION, STEMNESS AND MIGRATION Next, we examined the role of
SOX9 in CCA cell proliferation, stemness and migration. MTT analyses showed that knockdown of SOX9 expression significantly inhibited cell proliferation in four types of CCA cells (Fig. 5A).
In CC-SW-1 and EGI-1 cells, SOX9 inhibition significantly decreased the proportion of cells staying in G1 phase and increased those in G2/M phase (Fig. 5b). The results suggest that SOX9 is
required for maintaining CCA cell proliferation. Subsequently, we investigated the effects of SOX9 on stemness of CCA cells. We found that knockdown of SOX9 expression decreased EpCAM
expression at both RNA and protein levels in CC-SW-1 cells (Fig. 5c). In HCC, EpCAM is considered as a crucial factor in the maintenance of CSC-like features in cancer cells.24 To
investigate whether SOX9 is implicated to the CSC features of CCA cells, we performed tumour sphere formation assay, a widely recognised method to evaluate cancer stem cell self-renewal and
differentiation at the single-cell level in vitro.25 SOX9 knockdown significantly inhibited the capacity of tumour sphere formation in CC-SW-1 (Fig. 5d). In addition, we also investigated
the role of SOX9 in CCA cell migration. Transwell assay showed that knockdown of SOX9 expression significantly inhibited cell migration in CC-SW-1 cells (Fig. 5e). DISCUSSION The standard
treatment for advanced-stage iCCA is systemic chemotherapy with gemcitabine and cisplatin.7 However, the median OS time is <12 months.7 In patients treated with gemcitabine alone, the
survival time is <8 months,7 thus improving the sensitivity of cholangiocarcinoma cells to chemotherapy is a key to prolonging the survival of iCCA patients. In the current study, we
found that SOX9, the earliest cholangiocyte marker during embryonic liver development,11,12 plays a crucial role in iCCA cells’ resistance to chemotherapy. We examined the expression of SOX9
in 59 iCCA patients who received surgery. High expression of SOX9 in the nuclei of iCCA cancer cells was significantly associated with shorter survival time (_P_ = 0.0039). Of nine patients
treated with chemotherapy following surgery, the median survival time reached 62 months in six patients who had low levels of SOX9 expression, whereas survival time was only 22 months in
the three patients who had high SOX9 levels. Although the sample size was small in this study, the difference in survival time between both groups was significant (_P_ = 0.017). Among the
nine patients, eight received chemotherapy because of the recurrence of cancer. The five patients with low SOX9 levels survived between 16 and 34 months; however, the longest survival time
in three patients with high SOX9 expression was only 16 months. These results suggest that SOX9 expression correlates with cholangiocellular cancer cells’ response to chemotherapy. Further
in vitro studies provided the following mechanistic explanations of the observed differences:1 Microarray, qPCR and western blot analyses showed that disruption of SOX9 with siRNA
significantly decreased expression of genes/proteins associated with drug metabolism and multidrug resistance and increased the abundance of genes associated with p53 signalling pathway.2
Knockdown of SOX9 markedly inhibited gemcitabine-induced activation of CHK1, a key cell cycle checkpoint protein that coordinates the DNA damage response, and expression of multiple drug
resistance protein MRP4, and thus increased cancer cell apoptosis.3 Gemcitabine dose-dependently induced expression of SOX9, indicating that CCA cells increase SOX9 as a defensive mechanism
against treatment with chemotherapeutics such as gemcitabine. Figure 6 depicts the gemcitabine-induced loop in CCA cells. The role of SOX9 in the chemoresistance has been reported in
multiple tumours, for example, chondrosarcoma, breast cancer, glioblastoma, cervical cancer, gastric cancer and lung cancer.26,27,28,29,30,31 In cervical cancer cells, SOX9 was found to
increase cancer cell chemoresistance through inhibiting miR-130a.27 However, the detailed mechanisms of how SOX9 contributes to chemoresistance have not been clarified to date. Our
observation that SOX9 confers chemoresistance to cholangiocarcinoma through the activation of CHK1 and the expression of multiple drug resistance proteins might provide a light pointer for
further investigation of this aspect. The current study also investigated the role of SOX9 in CCA cells receiving cisplatin. In contrast to gemcitabine, knockdown of SOX9 did not impact the
efficiency of cisplatin. Administration of cisplatin did not alter expression of pChk1 in CCA cells. The discrepancy between the two compounds might be due to their different mechanisms of
action. Gemcitabine results in cell death mainly through the inhibition of DNA synthesis and the inhibition of enzymes relevant to deoxyribonucleotide metabolism.32 Thus, gemcitabine exerts
these actions through impacting multiple pathways, including regulating checkpoint kinases. Different from gemcitabine, cisplatin causes cell death through the formation of
[PtCl(guanine-DNA)(NH3)2]+, which inhibits DNA repair and activates apoptosis.33 The defensive effects of SOX9 for CCA cells are not limited to cancer cells facing chemotherapy. Knockdown of
SOX9 in both iCCA and eCCA cells remarkably inhibited the capacity of cancer cell proliferation and migration, decreased CSC stemness and increased apoptosis. These results provide an
explanation why SOX9 expression is associated with survival time of patients receiving chemotherapy, but also in those who are treated by surgery alone. On the other hand, it should be kept
in mind that these in vitro findings are not always consistent with the observation obtained in patients. For example, there was no correlation between the expression of SOX9 and cell
proliferation markers, for example, Ki67 and PCNA, in iCCA patients (data not shown). Given that there are multiple growth factors and proliferative signals governing cancer cell expansion
in patients with iCCA, it is not surprising that low levels of SOX9 expression alone does not have a significant impact on the proliferation of cancer cells. In addition, although there is a
crucial role of SOX9 in maintaining cell identity, we did not observe a significant correlation between SOX9 expression and histological differentiation in this cohort of iCCA patients
(data not shown). The result might have two explanations:1 SOX9 expression does not impact on differentiation of iCCA, and2 SOX9 might have a subtle influence on cancer cell differentiation,
but the association is too small to be detected in the currently small number of specimens. In diseased liver, high levels of SOX9 occur not only in CCA but also in HCC. However, in
contrast to cholangiocytes, normal hepatocytes do not express SOX9. Expression of SOX9 in HCC reflects a cancer stem cell/progenitor cell.18 Given that SOX9 is the earliest and dominant
phenotype marker of normal cholangiocytes, the induction of a cancer stem cell-like phenotype should not be attributed to the expression of SOX9. However, the current results do suggest that
SOX9 plays a role in the maintenance of cancer stem cell phenotypes. Like CCA patients, HCC patients with high levels of SOX9 had poor prognosis. As in CCA, SOX9 in HCC is implicated in
maintaining proliferation and self-renewal of cancer cells.18 In the future, it will be interesting to find out whether SOX9-dependent control of checkpoint protein activation may also play
a role in chemoresistance of HCC. Besides SOX9, this study analysed the association between CK19 expression and the clinical outcome of iCCA. CK19 is a classical marker for cholangiocytes.
It has been reported that CK19 contributes to the differentiation of iCCA from metastatic adenocarcinoma and is associated with the histological differentiation of iCCA.34 In the current
cohort of iCCA patients, CK19 expression was correlated with AJCC classification of iCCA, whereas it did not show any association with the survival of iCCA patients. Taken together, SOX9
expression is a sensitive marker that predicts the survival time of iCCA patients, particularly in those receiving chemotherapy. Our study demonstrates that SOX9 is a key transcription
factor that prevents iCCA cells from apoptosis when the cells are attacked by drugs such as gemcitabine. SOX9 exerts the observed effects on CCA cells, at least in part, through the
activation of Chk1 and upregulation of multidrug resistance genes. Limitations of this study are: (1) the low number of samples precludes more general conclusions; (2) it is not clear how
SOX9 exerts the observed effects on the expression of multidrug resistance genes; (3) we cannot conclude whether SOX9 expression has a similar effect in eCCA. Our study also enroled five
eCCA patients who demonstrate a similar biological behaviour as iCCA (data not shown). Results in eCCA cell line EGI-1 indicate that the SOX9 expression has similar effects as observed in
iCCA. Further investigation based on a large size of patient cohorts is required in the future. REFERENCES * Moeini, A., Sia, D., Bardeesy, N., Mazzaferro, V. & Llovet, J. M. Molecular
pathogenesis and targeted therapies for intrahepatic cholangiocarcinoma. _Clin. Cancer Res._ 22, 291–300 (2016). Article Google Scholar * Bridgewater, J. et al. Guidelines for the
diagnosis and management of intrahepatic cholangiocarcinoma. _J. Hepatol._ 60, 1268–1289 (2014). Article Google Scholar * Njei, B. Changing pattern of epidemiology in intrahepatic
cholangiocarcinoma. _Hepatology_ 60, 1107–1108 (2014). Article Google Scholar * Rizvi, S. & Gores, G. J. Pathogenesis, diagnosis, and management of cholangiocarcinoma.
_Gastroenterology_ 145, 1215–1229 (2013). Article CAS Google Scholar * Rizvi, S. & Gores, G. J. Emerging molecular therapeutic targets for cholangiocarcinoma. _J. Hepatol._ 67,
632–644 (2017). * DeOliveira, M. L. et al. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. _Ann. Surg._ 245, 755–762 (2007). Article Google Scholar
* Valle, J. et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. _N. Engl. J. Med._ 362, 1273–1281 (2010). Article CAS Google Scholar * Gordon, C. T. et al.
Long-range regulation at the SOX9 locus in development and disease. _J. Med. Genet._ 46, 649–656 (2009). Article CAS Google Scholar * Pritchett, J., Athwal, V., Roberts, N., Hanley, N. A.
& Hanley, K. P. Understanding the role of SOX9 in acquired diseases: lessons from development. _Trends Mol. Med._ 17, 166–174 (2011). Article CAS Google Scholar * Jo, A. et al. The
versatile functions of Sox9 in development, stem cells, and human diseases. _Genes Dis._ 1, 149–161 (2014). Article CAS Google Scholar * Antoniou, A. et al. Intrahepatic bile ducts
develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. _Gastroenterology_ 136, 2325–2333 (2009). Article Google Scholar * Si-Tayeb, K., Lemaigre, F.
P. & Duncan, S. A. Organogenesis and development of the liver. _Dev. Cell_ 18, 175–189 (2010). Article CAS Google Scholar * Carpino, G. et al. Biliary tree stem/progenitor cells in
glands of extrahepatic and intraheptic bile ducts: an anatomical in situ study yielding evidence of maturational lineages. _J. Anat._ 220, 186–199 (2012). Article Google Scholar *
Kawaguchi, Y. Sox9 and programming of liver and pancreatic progenitors. _J. Clin. Invest._ 123, 1881–1886 (2013). Article CAS Google Scholar * Jors, S. et al. Lineage fate of ductular
reactions in liver injury and carcinogenesis. _J. Clin. Invest._ 125, 2445–2457 (2015). Article Google Scholar * Cardinale, V. et al. Multipotent stem/progenitor cells in human biliary
tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. _Hepatology_ 54, 2159–2172 (2011). Article CAS Google Scholar * Leung, C. O. et al. Sox9 confers stemness properties
in hepatocellular carcinoma through Frizzled-7 mediated Wnt/beta-catenin signaling. _Oncotarget_ 7, 29371–29386 (2016). PubMed PubMed Central Google Scholar * Liu, C. et al. Sox9
regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. _Hepatology_ 64, 117–129 (2016). Article CAS Google
Scholar * Matsushima, H. et al. Sox9 expression in carcinogenesis and its clinical significance in intrahepatic cholangiocarcinoma. _Dig. Liver Dis._ 47, 1067–1075 (2015). Article CAS
Google Scholar * Weng, H. L. et al. The etiology of liver damage imparts cytokines transforming growth factor beta1 or interleukin-13 as driving forces in fibrogenesis. _Hepatology_ 50,
230–243 (2009). Article CAS Google Scholar * Meyer, C., Liu, Y., Kaul, A., Peipe, I. & Dooley, S. Caveolin-1 abrogates TGF-beta mediated hepatocyte apoptosis. _Cell Death Dis._ 4,
e466 (2013). Article CAS Google Scholar * Dai, M. et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. _Nucleic Acids Res._ 33, e175
(2005). Article Google Scholar * Otto, T. & Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. _Nat. Rev. Cancer_ 17, 93–115 (2017). Article CAS Google Scholar
* Yamashita, T. et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. _Gastroenterology_ 136, 1012–1024 (2009). Article CAS
Google Scholar * Pastrana, E., Silva-Vargas, V. & Doetsch, F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. _Cell Stem Cell_ 8, 486–498 (2011).
Article CAS Google Scholar * Chen, W. et al. MALAT1-miR-101-SOX9 feedback loop modulates the chemo-resistance of lung cancer cell to DDP via Wnt signaling pathway. _Oncotarget_ 8,
94317–94329 (2017). PubMed PubMed Central Google Scholar * Feng, C. et al. SOX9/miR-130a/CTR1 axis modulates DDP-resistance of cervical cancer cell. _Cell Cycle_ 17, 448–458 (2018).
Article CAS Google Scholar * Garros-Regulez, L. et al. mTOR inhibition decreases SOX2-SOX9 mediated glioma stem cell activity and temozolomide resistance. _Expert Opin. Ther. Targets_ 20,
393–405 (2016). Article CAS Google Scholar * Liang, Z., Bian, X. & Shim, H. Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer.
_Biochem. Biophys. Res. Commun._ 477, 461–466 (2016). Article CAS Google Scholar * Riemenschnitter, C., Teleki, I., Tischler, V., Guo, W. & Varga, Z. Stability and prognostic value of
Slug, Sox9 and Sox10 expression in breast cancers treated with neoadjuvant chemotherapy. _+_ 2, 695 (2013). PubMed PubMed Central Google Scholar * Wang, J. et al. Upregulation of
microRNA-524-5p enhances the cisplatin sensitivity of gastric cancer cells by modulating proliferation and metastasis via targeting SOX9. _Oncotarget_ 8, 574–582 (2017). PubMed Google
Scholar * de Sousa Cavalcante, L. & Monteiro, G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. _Eur. J. Pharmacol._
741, 8–16 (2014). Article Google Scholar * Wang, D. & Lippard, S. J. Cellular processing of platinum anticancer drugs. _Nat. Rev. Drug Discov._ 4, 307–320 (2005). Article CAS Google
Scholar * Shimonishi, T., Miyazaki, K. & Nakanuma, Y. Cytokeratin profile relates to histological subtypes and intrahepatic location of intrahepatic cholangiocarcinoma and primary sites
of metastatic adenocarcinoma of liver. _Histopathology_ 37, 55–63 (2000). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We are grateful to Drs. N. Kobayashi, F. Rückert
and L.G. Lu for providing normal cholangiocyte and CCA cell lines and gemcitabine. We thank the core facilities of Biosit and Biogenouest (CRB Santé BB-0033-00056 and H2P2) for TMA
production. The study was supported by Chinese-German Cooperation Group project, grant number: GZ 1263 (H.-L.W., M.P.A.E., S.D.), Deutsche Forschungsgemeinschaft Do373/13-1 (S.D.), Ligue
Contre le Cancer – Comité départemental des Côtes-d’Armor (C.C.), and Novartis Oncology (L.S.). AUTHOR CONTRIBUTIONS Conception, hypothesis and experimental design: X.Y., S.D. and H.-L.W.
Drafting the article: X.Y. and H.-L.W. Collecting patients’ data and sampling: J.L., L.S., D.B., C.C. and H.-L.W. Immunohistochemistry and microarray analysis: X.Y., C.D.L.T., L.S., D.B. and
C.C. Reviewing and editing the article critically: X.Y., J.L., C.C., L.S., D.B., D.L.T.C., R.L., N.G., M.P.A.E., S.D. and H.-L.W. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of
Medicine II, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany Xiaodong Yuan, Tao Lin, Matthias P. A. Ebert, Steven Dooley & Hong-Lei Weng * Department of General
Visceral Transplantation, University Hospital Tübingen, Tübingen, Germany Jun Li * Inserm, Inra, University Rennes, UMR 1241, Nutrition Metabolisms and Cancer (NuMeCan), Service de Chirurgie
Hépatobiliaire et Digestive, Rennes, France Cédric Coulouarn, Laurent Sulpice & Damien Bergeat * Medical Research Center, Medical Faculty of Mannheim, University of Heidelberg,
Mannheim, Germany Carolina De La Torre & Norbert Gretz * Department of Medicine II, Saarland University Medical Center, Saarland University, Homburg, Germany Roman Liebe Authors *
Xiaodong Yuan View author publications You can also search for this author inPubMed Google Scholar * Jun Li View author publications You can also search for this author inPubMed Google
Scholar * Cédric Coulouarn View author publications You can also search for this author inPubMed Google Scholar * Tao Lin View author publications You can also search for this author
inPubMed Google Scholar * Laurent Sulpice View author publications You can also search for this author inPubMed Google Scholar * Damien Bergeat View author publications You can also search
for this author inPubMed Google Scholar * Carolina De La Torre View author publications You can also search for this author inPubMed Google Scholar * Roman Liebe View author publications You
can also search for this author inPubMed Google Scholar * Norbert Gretz View author publications You can also search for this author inPubMed Google Scholar * Matthias P. A. Ebert View
author publications You can also search for this author inPubMed Google Scholar * Steven Dooley View author publications You can also search for this author inPubMed Google Scholar *
Hong-Lei Weng View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Steven Dooley or Hong-Lei Weng. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing interests. NOTE This work is published under the standard license to publish agreement. After 12 months the work will become freely
available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0). ADDITIONAL INFORMATION Note: This work is published under the standard license to
publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0). ELECTRONIC
SUPPLEMENTARY MATERIAL AUTHOR CHANGE LETTER SUPPLEMENTARY FIGURE 1 SUPPLEMENTARY FIGURE 2 SUPPLEMENTARY FIGURE 3 SUPPLEMENTARY TABLE 1 SUPPLEMENTARY TABLE 2 SUPPLEMENTARY TABLE 3
SUPPLEMENTARY TABLE 4 SUPPLEMENTARY TABLE 5 RIGHTS AND PERMISSIONS This article is distributed under the terms of the Creative Commons Attribution 4.0 International License
(http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s)
and the source, provide a link to the Creative Commons license, and indicate if changes were made. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Yuan, X., Li, J., Coulouarn,
C. _et al._ SOX9 expression decreases survival of patients with intrahepatic cholangiocarcinoma by conferring chemoresistance. _Br J Cancer_ 119, 1358–1366 (2018).
https://doi.org/10.1038/s41416-018-0338-9 Download citation * Received: 23 June 2018 * Revised: 20 October 2018 * Accepted: 24 October 2018 * Published: 13 November 2018 * Issue Date: 27
November 2018 * DOI: https://doi.org/10.1038/s41416-018-0338-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
