Subtyping-based platform guides precision medicine for heavily pretreated metastatic triple-negative breast cancer: the future phase ii umbrella clinical trial
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Triple-negative breast cancer (TNBC) is a heterogeneous disease and lacks effective treatment. Our previous study classified TNBCs into four subtypes with putative therapeutic
targets. Here, we report the final results of FUTURE, a phase II umbrella trial designed to explore whether the subtyping-based strategy may improve the outcomes in metastatic TNBC patients.
A total of 141 patients with a median of three previous lines of therapies in the metastatic setting were enrolled in seven parallel arms. Confirmed objective responses were achieved in 42
patients (29.8%; 95% confidence interval [CI], 22.4–38.1). The median values of progression-free survival and overall survival were 3.4 (95% CI: 2.7–4.2) and 10.7 (95% CI: 9.1–12.3) months,
respectively. Given Bayesian predictive probability, efficacy boundaries were achieved in four arms. Furthermore, integrated genomic and clinicopathological profiling illustrated
associations of clinical and genomic parameters with treatment efficacy, and the efficacy of novel antibody–drug conjugates was explored in preclinical TNBC models of subtypes for which
treatment was futile. In general, the FUTURE strategy recruits patients efficiently and provides promising efficacy with manageable toxicities, outlining a direction for further clinical
exploration. SIMILAR CONTENT BEING VIEWED BY OTHERS MOLECULAR SUBTYPING AND GENOMIC PROFILING EXPAND PRECISION MEDICINE IN REFRACTORY METASTATIC TRIPLE-NEGATIVE BREAST CANCER: THE FUTURE
TRIAL Article Open access 27 July 2020 PRACTICAL CLASSIFICATION OF TRIPLE-NEGATIVE BREAST CANCER: INTRATUMORAL HETEROGENEITY, MECHANISMS OF DRUG RESISTANCE, AND NOVEL THERAPIES Article Open
access 16 October 2020 GENOMIC CHARACTERIZATION REVEALS DISTINCT MUTATIONAL LANDSCAPES AND THERAPEUTIC IMPLICATIONS BETWEEN DIFFERENT MOLECULAR SUBTYPES OF TRIPLE-NEGATIVE BREAST CANCER
Article Open access 29 May 2024 INTRODUCTION Triple-negative breast cancer (TNBC) is pathologically defined as a subgroup of breast cancers that lacks estrogen receptor (ER), progesterone
receptor (PR) and human epidermal growth factor receptor 2 (HER2) expression;1,2 this precludes the use of targeted therapies, and the most available systemic treatment option is
chemotherapy. Accounting for approximately 15% of invasive breast cancers, TNBC is associated with a high risk of early recurrence and poor patient outcome. The low response rate (5%–10%) of
TNBC to standard chemotherapy in the later-line settings highlights the need for advances in therapeutic options.3,4,5 In recent years, immunotherapy has emerged as a breakthrough for
treating TNBC,6,7,8 while PARP inhibitors have provided a significant benefit for patients carrying _BRCA_ germline mutations.9,10 However, these treatments are far from satisfactory for
metastatic TNBC due to the lack of evidence supporting immunotherapy in later-line treatment11 and the low prevalence of _BRCA_ germline mutations.12 Recently, sacituzumab govitecan and
fam-trastuzumab deruxtecan-nxki (DS-8201a; T-DXd; tradename Enhertu [Daiichi Sankyo]), two novel antibody–drug conjugates (ADCs), have been successively granted regular approval by the U.S.
Food and Drug Administration for patients with metastatic TNBC and patients with metastatic HER2-low breast cancer, respectively.4,13 The identification of specific DNA alterations for
available targeted therapies has also opened the door for genome-driven cancer treatment,14,15,16,17 but only a small fraction of TNBCs have targetable mutations.18,19 Owing to the overall
poor prognosis and the complexity of molecular features of TNBC, there is an ongoing need to find effective therapeutic matches. Studies describing targeted therapies for TNBC have laid the
groundwork for precision medicine.20,21,22 However, those studies mainly focused on specific targets, ignoring the intrinsic subtypes of TNBCs and limiting the enrollment of TNBC patients
without druggable targets, which reduced their clinical applicability. Our previous study presented multi-omic profiling of 465 Chinese TNBCs and classified them into four subtypes, namely,
luminal androgen receptor (LAR), immunomodulatory (IM), basal-like immune-suppressed (BLIS) and mesenchymal-like (MES).18 Putative treatment options were then identified for each subtype,
allowing for a broader population to achieve precision treatment. Building on that advancement, we developed an immunohistochemistry (IHC)-based classification approach, which simplified and
increased the clinical utility of the subtyping system.23 Here, we aimed to assess the efficacy and safety of molecular subtyping and genomic sequencing-guided precision therapy for heavily
pretreated metastatic TNBC. Our study recruited metastatic TNBC patients who were resistant to the most common chemotherapeutic agents used in breast cancer treatment.1 In the interim
analysis, the outcomes were favorable, with an objective response rate (ORR) of 29.0% for 69 enrolled patients.24 Here, we report the final clinical efficacy (including survival data for the
first time), safety profile, biomarker analysis, and exploration of optimized regimens of the Fudan University Shanghai Cancer Center TNBC umbrella (FUTURE) trial. RESULTS PATIENT
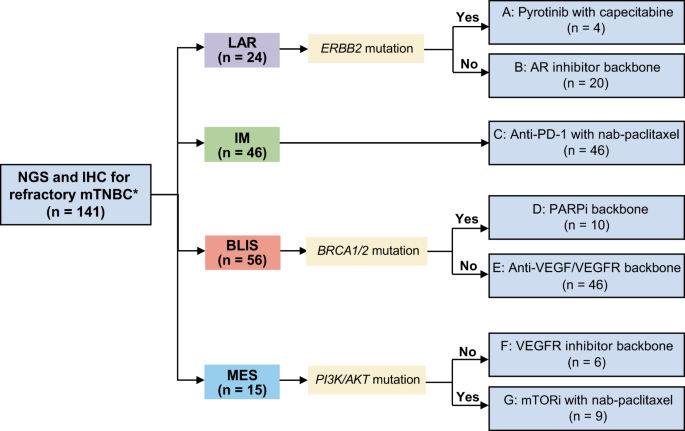
CHARACTERISTICS Between October 18, 2018, and February 11, 2022, 141 patients were enrolled (Fig. 1). All patients were heavily pretreated (median of 3 previous lines of antitumor regimens
in the metastatic setting [range, 1–8]), and most of them had received taxane (99.3%), anthracycline (92.9%), platinum (93.6%), vinorelbine (80.9%), capecitabine (87.9%) and gemcitabine
(70.2%). The baseline characteristics are summarized in Table 1. The presence of germline _BRCA1/2_ mutations and the expression of PD-L1 are shown in Supplementary information, Table S1,
Table S2, respectively. The median age was 50 years (range, 23–74). Seventy-one (50.4%) patients had three or more metastatic sites, 69 (48.9%) patients had lung metastasis, and 43 (30.5%)
patients had liver metastasis. At the data cutoff (March 31, 2022), the median follow-up time was 18.3 months (95% CI: 16.7–19.9). Nine patients continued to receive treatment in the trial.
Detailed information and reasons for discontinuation are shown in Supplementary information, Fig. S1. PRIMARY AND SECONDARY OUTCOMES OF THE WHOLE COHORT Patients were enrolled into one of
the following arms based on their TNBC subtypes and genomic features: (A) pyrotinib with capecitabine, (B) androgen receptor inhibitor backbone therapy, (C) anti-PD-1 with nab-paclitaxel,
(D) PARP inhibitor backbone therapy, (E) anti-VEGF/VEGFR backbone therapy, (F) VEGFR inhibitor backbone therapy, and (G) mTOR inhibitor with nab-paclitaxel (Fig. 1). A total of 112 of the
141 enrolled patients underwent at least one postbaseline assessment (Supplementary information, Fig. S2), and the reasons for the remaining 29 patients not undergoing postbaseline
assessments are listed in Supplementary information Table S3. In general, an objective response (complete response [CR] and partial response [PR]) was achieved in 42 (29.8%; 95% CI:
22.4–38.1) patients (Fig. 2a, b and Table 2), with a median time to response of 1.8 months and a median duration of response of 4.9 months. At the data cutoff, nine patients had long-term
responses for more than 12 (range, 13.6–19.7) months (Fig. 2c). Disease control was achieved in 68 (48.2%; 95% CI: 39.7–56.8) patients (Fig. 2d). Moreover, we formally disclosed the survival
data of the FUTURE trial. The median progression-free survival (PFS) was 3.4 (95% CI: 2.7–4.2) months; the estimated probability of PFS was 26.5% (95% CI: 22.4–30.6) at 6 months and 12.4%
(95% CI: 9.2–15.6) at 12 months (Fig. 2e). At the data cutoff, 63.8% (90/141) of overall survival (OS) events were recorded. The median OS was 10.7 (95% CI: 9.1–12.3) months; the estimated
probability of survival was 66.8% (95% CI: 62.7–70.9) at 6 months and 44.2% (95% CI: 39.7–48.7) at 12 months (Fig. 2f). Detailed survival events and censoring proportions of each arm are
described in Supplementary information, Table S4. ARMS ACHIEVING PRESPECIFIED EFFICACY BOUNDARIES: A, C, E, G According to the study design, arms A, C, E, and G reached the efficacy
boundaries based on Bayesian prediction probability (see the “Bayesian prediction probability” part of the Materials and Methods section; more details in Supplementary information, Data
S1).25 As shown in the waterfall plot, which depicted tumor responses in patients of arms A, C, E, and G with at least one response assessment, a reduction in target lesions was achieved in
61.0% (64/105) of patients (Fig. 3a). Examples of patients achieving objective responses are shown in Fig. 3b. Four patients who had the LAR subtype with _ERBB2_ mutation identified by
next-generation sequencing (NGS) were enrolled in arm A, and confirmed objective responses were achieved in 3 patients, with a median PFS of 3.4 (95% CI: 0–7.3) months and a median OS of
16.7 (95% CI: 0–35.3) months. Arm A was terminated early due to its high response rate and the low prevalence of _ERBB2_ alteration. Forty-six patients who had the IM subtype were enrolled
in arm C and received camrelizumab plus nab-paclitaxel. Arm C was also terminated early as it reached the prespecified sample size (_n_ = 46, more details in Supplementary information, Data
S1) and tumor responses are shown in Supplementary information, Fig. S3a. In general, arm C achieved an ORR of 43.5% (95% CI: 28.9–58.9; Fig. 2a, b), with a median PFS of 4.6 (95% CI:
3.4–5.9) months and a median OS of 16.1 (95% CI: 11.7–20.5) months (Supplementary information, Table S4). Of the 20 patients meeting confirmed objective responses, the median duration of
response was 8.6 (range 1.2–19.7) months (Supplementary information, Fig. S3b, c). Forty-six patients who had the BLIS subtype without germline _BRCA1/2_ mutation were enrolled in arm E to
receive anti-VEGF/VEGFR backbone therapy. Thirteen patients achieved confirmed objective responses, with 1 CR and 12 PRs (Supplementary information, Fig. S3d). The ORR was 28.3% (95% CI:
16.0–43.5; Fig. 2a, b), with a median PFS of 3.4 (95% CI: 1.7–5.0) months and a median OS of 10.1 (95% CI: 3.8–16.3) months (Supplementary information, Table S4). Among the 13 patients
meeting confirmed objective responses, the median duration of response was 4.2 (range 0.9–15.3) months (Supplementary information, Fig. S3e, f). Nine patients who had the MES subtype with
_PI3K/AKT_ mutation were enrolled in arm G, and confirmed objective responses were achieved in 3 patients, with a median PFS of 3.0 (95% CI: 2.4–3.6) months and a median OS of 4.5 (95% CI:
2.4–6.6) months (Supplementary information, Table S4). ARMS NOT ACHIEVING PRESPECIFIED EFFICACY BOUNDARIES: B, D, F According to the study design, arms B, D, and F did not reach the efficacy
boundaries (see the “Bayesian prediction probability” part of the Materials and Methods section; more details in Supplementary information, Data S1). Twenty patients who had the LAR subtype
without _ERBB2_ mutations were enrolled in arm B and received an AR inhibitor as the backbone of their treatment. Despite multiple adjustments to the combination regimen of AR inhibitors
(more details in Supplementary information, Data S1), none of the patients achieved confirmed objective responses. Arms D and F remained unclosed according to the study protocol until data
cutoff. Ten patients who had the BLIS subtype with _BRCA_ germline mutations were enrolled in arm D, and confirmed objective responses were achieved in two patients. Six patients who had the
MES subtype without _PI3K/AKT_ mutations were enrolled in arm F, and a confirmed objective response was achieved in only one patient. Detailed survival data for arms D and F are shown in
Table 2 and Supplementary information, Table S4. SAFETY Safety data were consistent with the known safety profiles of relevant drugs.24,26 No grade 5 treatment-related adverse events (TRAEs)
were reported. The most common grade 3–4 adverse events (AEs) were leukopenia (15.6%), neutropenia (14.9%), anemia (12.1%), and thrombocytopenia (9.9%), while the most common grade 3–4
nonhematologic events were hypertension (5.0%) and proteinuria (2.8%). AEs of any grade occurring in more than 10% of patients and grade 3–4 AEs occurring in ≥ 1 patient are summarized in
Table 3. The treatment discontinuation (due to AE) rate was 8.5%, and the dose reduction and/or delay rate was 23.4%. The number of AEs of any grade and grade 3–4 occurring in each arm are
shown in Supplementary information, Table S5. POST HOC BIOMARKER ANALYSIS We then explored clinical features and genomic events associated with treatment response and tested the efficacy of
novel ADCs in different subtypes of TNBCs to further inform precision oncology (Fig. 4a). We first evaluated the ORR in a variety of clinical subgroups in the FUTURE trial. Patients with
more than 3 different organ metastases had a significantly lower ORR (17.1% [95% CI: 7.2–32.1] vs 35.0% [95% CI: 25.7–45.2]), while those over 50 years of age at diagnosis had a higher ORR
(39.4% [95% CI: 28.0–51.7] vs 20.0% [95% CI: 11.4–31.3]) (Supplementary information, Table S6). Genomic analysis included 129 (91.5%) patients with Fudan University Shanghai Cancer Center
(FUSCC) NGS sequencing. The most prevalent somatic variations were _TP53_ (78%), _PIK3CA_ (25%) and _PKD1_ (11%) (Fig. 4b). High mutation frequencies were observed in the genome integrity
(83%), PI3K signaling (44%) and RTK signaling (25%) pathways (Supplementary information, Fig. S4). We then used univariate Cox regression analyses to explore the predictive value of frequent
somatic mutations (≥ 5%) for PFS in each arm. Interestingly, a reduced clinical benefit of immunotherapy (arm C) was observed in patients with _PDGFRB_, _UNC13D_ or _TSC2_ mutations;
patients with _EPHB1_ mutation had shorter PFS in arm E (Fig. 4c). Additionally, we analyzed the genomic characteristics of patients treated with everolimus for _PIK3CA_ mutation (B and G
arms). Despite the similar _PI3KCA_ mutation site (seven with H1047R and one with H1047L), the favorable outcome of everolimus was observed only in arm G (Supplementary information, Fig.
S5), further suggesting the need for a subtyping-based precision treatment strategy. ADCS PROVIDE GREAT EFFICACY IN BLIS AND LAR SUBTYPES Considering the poor outcomes in BLIS and LAR
subtype patients, we then tried to explore new treatment strategies for these patients. ADCs have shown strong antitumor activity in solid tumors, especially breast cancer.27 For breast
cancer, anti-HER2 ADCs and anti-Trop-2 ADCs are promising.4,13 In our multi-omic data from a TNBC patient cohort (_n_ = 360),18 we observed that the expression of _ERBB2_ (encoding HER2
protein) was higher in the LAR subtype than in other subtypes, both at the protein (_P_ < 0.001, Fig. 5a) and mRNA levels (_P_ < 0.001, Fig. 5b), suggesting that patients diagnosed
with the TNBC LAR subtype might benefit from anti-HER2 ADCs. Interestingly, in LAR and BLIS subtype patients, the mRNA expression of _TACSTD2_ (encoding Trop-2 protein) was higher than that
in IM and MES subtype patients, suggesting that the LAR and BLIS subtypes might be sensitive to anti-Trop-2 ADC (_P_ < 0.001, Fig. 5c). Subsequently, we investigated the efficacy of these
two ADCs in TNBC cell lines and patient-derived organoids (PDOs) grouped by subtype (Fig. 5d–g; Supplementary information, Fig. S6a). Notably, both models showed that the LAR subtype had
lower IC50 and viability in response to anti-HER2 ADC RC48 (Fig. 5d, f). In addition, tumors of LAR and BLIS subtypes had better responses to anti-Trop-2 ADC sacituzumab govitecan (SG) (Fig.
5e, g). Moreover, the expression levels of HER2 and Trop-2 in the TNBC cell lines and PDOs correlated with the ADCs efficacy (Supplementary information, Figs. S6b, S7). Collectively, ADCs
showed great efficacy in BLIS and LAR subtypes, holding promise for future design of precision strategies. DISCUSSION In this prospective umbrella trial, we evaluated the feasibility and
clinical utility of the subtyping-based precision strategy in heavily pretreated metastatic TNBC patients. Our work established a subtyping platform to navigate the precision treatment of
TNBC based on the recognition of molecular characteristics instead of genomic alterations. The main purpose of this work was to highlight the superiority of the platform, not the superiority
of a specific drug or regimen, as future advances in drug development could supplant the leading drugs. Notable accomplishments in this study include the following: (1) we demonstrated that
TNBC subtyping combined with NGS was clinically feasible for matching and enrolling patients, with new biomarker-driven treatment arms being introduced and conducted simultaneously after
previous arms reached a futility or efficacy boundary; (2) promising outcomes were confirmed in a subtype and genomic characteristics dual-directed therapeutic strategy, and these outcomes
can be translated into long-term survival benefit; (3) integrated genomic and clinicopathological profiling illustrated associations of clinical and genomic parameters with treatment
efficacy, and for arms with unsatisfactory response, novel ADCs were tested, providing clues for further exploration. The FUTURE study recruited 141 TNBC patients. Unlike most umbrella
trials focusing on specific therapies,14,15,28 this study was mainly driven by molecular subtyping rather than single gene alterations. Therefore, because of treatment allocation according
to molecular subtype, FUTURE allowed enrollment of more screened patients (93.4% [141/151] compared with a 10%–20% enrollment rate in most biomarker-driven studies), which was critical for
mTNBC patients with limited treatment options after progression on multiple lines of chemotherapy. In addition, Bayesian predictive probability was adopted to make adequacy of sample size of
each arm more flexible,25 allowing the potential efficacy of the drug combination to be tested quickly and efficiently, especially in the arms with relatively low enrollment rates. In
addition, patients enrolled in this study were heavily pretreated and chemotherapy resistant with a short disease-free interval and duration of first-line therapy. In this study, the ORR
reached nearly 30%, the median PFS reached 3.4 months, and the median OS reached 10.7 months, all of which were more favorable than the outcomes of traditional chemotherapy in the heavily
pretreated patients of the TNBC population (ORR of 5%, median PFS of 1.7 months, and median OS of 6.7 months).4 Among the reported 7 arms, arms A, C, E, and G reached an efficacy boundary
and arm B reached a futility boundary, while the data from arms D and F were immature due to the low proportion of these 2 specific subgroups. The imbalance of patient numbers between
different arms may reflect the natural distribution of TNBC molecular subtypes in the metastatic setting as arms D and F remained open during the whole recruitment. Arms A, C, E, and G
demonstrated promising outcomes. For arm C, the results for PD-1 blockade plus chemotherapy in the IM subtype showed the highest ORR reported in a prospective trial conducted with heavily
pretreated metastatic TNBC patients. A highlight of arm C was the usage of CD8 to define the IM subtype and match “immune-hot” tumors, validating the initial hypothesis that we proposed.18
Arm C achieved an ORR of 43.5%, with a median PFS of 4.6 months and a median OS of 16.1 months. In addition, the favorable response was remarkably durable, with a median duration of 8.6
months. This was also validated in the FUTURE-C-Plus trial, where the camrelizumab backbone regimen achieved a confirmed ORR of 81.3% and a median PFS of 13.6 months in the first-line
treatment of CD8+ advanced TNBC patients.29,30 The subsequent randomized controlled phase III study (NCT05134194) is ongoing. BLIS is characterized by high expression of the VEGF signature,
which is associated with tumor angiogenesis and poor prognosis.18 In patients with the BLIS subtype without _BRCA_ germline mutation in arm E, we assessed the effect of anti-VEGF/VEGFR
therapy. A confirmed ORR of nearly 30% was achieved, which was higher than previously reported results in heavily pretreated TNBC patients.31 These findings suggested that anti-VEGF/VEGFR
therapy showed preliminary efficacy in _BRCA_ wild-type BLIS tumors, and it warrants further exploitation in _BRCA_-mutated patients. Bevacizumab or low-dose apatinib combined with VP-16 may
be more tolerated than apatinib 500 mg. Interestingly, arms A and G showed promising outcomes in a small sample size. In rare instances (2%–4%), patients with metastatic breast cancer have
_ERBB2_ mutations but are HER2-negative according to clinical guidelines.32 In patients with LAR subtypes, _ERBB2_ mutations were enriched.18 Arm A achieved a confirmed ORR of 75% after
receiving capecitabine plus pyrotinib. This arm suggested the potential of anti-HER2 therapy in tumors harboring _HER2_ mutations. Similarly, the SUMMIT study demonstrated that neratinib
combined with trastuzumab showed good antitumor activity in patients with _ERBB2_-mutated TNBC after previous multiline therapy, with an ORR of 33.3% and a median PFS of 6.2 months.33 A
total of 9 MES patients with _PI3K/AKT_ mutations were enrolled in arm G, and 3 of them achieved confirmed PRs. Similarly, the PAKT trial and the LOTUS trial showed that the addition of the
AKT inhibitor capivasertib or ipatasertib to first-line paclitaxel therapy resulted in significantly longer PFS, with more pronounced benefits in _PIK3CA/AKT1/PTEN_-altered tumors.34,35
Conversely, IPATunity130 failed to repeat the benefit in HER2-negative patients.36 Overall, these two arms proved the utility of FUTURE to evaluate drug activity in patients harboring rare
genomic aberrations in the context of molecular subtyping. By comparison, treatment efficacy was unsatisfactory in arms B, D, and F. For BLIS with _BRCA_ germline mutation, arm D tested the
efficacy of PARP inhibitors plus famitinib, but only 2 patients responded, which might be attributable to prior usage of platinum. All patients in arm D had previously undergone treatment
with platinum agents, and 8 of them had progressed during platinum treatment. Such patients were deemed resistant to PARP inhibitors and were excluded from clinical trials evaluating the
efficacy of PARP inhibitors.9,10 Moreover, we experimentally demonstrated that anti-Trop-2 ADC had a strong inhibitory effect in the BLIS subtype, probably due to higher _TACSTD2_ expression
in BLIS. Hence, anti-Trop-2 ADC might be promising in the treatment of BLIS subtype patients.37,38 Meanwhile, although previous studies suggested that the LAR subtype was enriched with
Chr9p21 loss, _CDKN2A_ losses/deletions18 and somatic mutations in the PI3K signaling pathway,39 the overall efficacy of arm B was disappointing, with no patient responding to AR inhibitors
with CDK4/6 inhibitors or mTOR inhibitors and with a median PFS of 1.9 months. In a recent study, bicalutamide plus abiraterone achieved a 19% clinical benefit rate in AR-positive advanced
TNBC.40,41 This inconsistency may be due to tumor evolution and patient selection after multiline chemotherapy in our study; another possibility is that AR may only be a biomarker rather
than a therapeutic target. Interestingly, we observed that the LAR subtype exhibited relatively higher _ERBB2_ and _TACSTD2_ expression. In vitro experiments confirmed that anti-HER2 and
anti-Trop-2 ADCs had strong inhibitory effects in cell lines and organoids of LAR subtype patients. Therefore, these drugs may be promising in treating LAR subtype patients and should be
tested in further studies. Finally, only six patients who had the MES subtype and no _PI3K/AKT_ mutations were enrolled in arm F, which does not provide sufficient grounds for a conclusion
at the current stage. FUTURE is a pilot study designed to generate clues for clinical practice. Owing to the design of this umbrella trial, the number of patients is relatively small in some
arms and unbalanced between arms, which has been discussed above. Notwithstanding the relatively limited sample size, this study offers valuable insights into tailoring and bolstering
precision therapy through prospective molecular subtyping selection. Next, direct comparison with other chemotherapy approaches was not available in this trial because of its noncomparative
design and the lack of accessible treatment options for the heavily pretreated patients of this population. Our randomized controlled umbrella FUTURE-SUPER trial (NCT04395989) is currently
in progress. Finally, although PFS was not as satisfactory as sacituzumab govitecan, the primary endpoints ORR and OS were both comparable.3 Likewise, as the first step in tailoring
treatment contingent on molecular subtyping in metastatic TNBC, the “FUTURE” strategy may constitute a platform to test the possibility of combining novel drugs within the framework of
subtyping hereafter. Collectively, the subtyping-based and genomic sequencing-guided strategy promotes promising efficacy with manageable toxicity in patients with heavily pretreated
metastatic TNBC. As a dynamic and ongoing platform for novel targeted regimens, FUTURE can enable efficient testing of potential new drug–biomarker combinations in the context of subtyping,
generating clues for further validation in expansion trials. MATERIALS AND METHODS STUDY DESIGN AND PARTICIPANTS FUTURE is a phase II, open-label, multicenter, umbrella trial evaluating the
efficacy and safety of multiple precision treatments based on molecular subtype and tumor characteristics in patients with heavily pretreated metastatic TNBCs. Eligibility criteria included
the following: (1) female patients diagnosed with metastatic breast carcinoma with an ER-negative, PR-negative, and HER2-negative phenotype (the IHC cutoff for ER/PR-negative status was less
than 1% staining in nuclei, and HER2-negative status was defined as a score of 0 or 1 by IHC analysis or the absence of ERBB2 amplification by fluorescence in situ hybridization with an IHC
score); (2) central pathologic examination of tumor specimens performed by the Department of Pathology at FUSCC; (3) an Eastern Cooperative Oncology Group (ECOG) performance status of 0 to
2; (4) at least one measurable lesion according to Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1; and (5) adequate hematologic function, hepatic function, and renal
function. Patients with uncontrolled brain metastasis were excluded from enrollment. Full eligibility criteria are provided in the study protocol in Supplementary information, Data S1. The
trial was conducted in accordance with the Declaration of Helsinki and the Good Clinical Practice guidelines of the International Conference on Harmonization. The study protocol was approved
by the institutional review board and ethics committee of FUSCC. The ethics committee reference number was 1807188-16. All patients provided written informed consent to participate in this
study. PROCEDURES Patients who had heavily pretreated metastatic TNBCs and experienced disease progression during or following almost all standard chemotherapies (anthracycline, taxane,
platinum, vinorelbine, capecitabine, and gemcitabine) were screened in four centers, including FUSCC, Beijing Cancer Hospital, Liaoning Cancer Hospital and Institute and the First Hospital
of Jilin University. Tumor biopsies were obtained to allow IHC staining (AR, CD8 and FOXC1) and NGS (Supplementary information, Table S7)23 in order to classify the tumors into four subtypes
(LAR, IM, BLIS, MES),24 and each patient was then enrolled into one of seven arms: (A) pyrotinib (HER1/HER2/HER4 inhibitor) 400 mg orally once daily continuously plus capecitabine 1000
mg/m2 orally twice daily from day 1 to day 14 on a 21-day cycle for LAR subtype with _ERBB2_ somatic mutation or amplification; (B) AR inhibitor (SHR3680) 240 mg orally once daily backbone
therapy for LAR subtype without _ERBB2_ somatic mutation or amplification; (C) anti-PD-1 antibody (SHR1210) 200 mg intravenously once every 2 weeks with nab-paclitaxel 100 mg/m2
intravenously on days 1, 8, and 15 in a 28-day cycle for IM subtype; (D) PARP inhibitor (SH3162) 150 mg orally twice daily plus VEGFR inhibitor (famitinib) 20 mg orally once daily for BLIS
subtype with _BRCA1/2_ germline mutation; (E) anti-VEGF/VEGFR backbone therapy for BLIS subtype without _BRCA1/2_ germline mutation; (F) VEGFR inhibitor (famitinib) orally once daily
continuously plus etoposide (VP-16) 50 mg orally once daily from day 1 to day 14 in a 21-day cycle for MES subtype without _PI3K/AKT_ mutation; (G) mTOR inhibitor (everolimus) 10 mg orally
once daily continuously with nab-paclitaxel by intravenous 100 mg/m2 on days 1, 8, and 15 in a 28-day cycle for MES subtype with _PI3K/AKT_ mutation. Details are provided in Table 5 of
Supplementary information, Data S1. BAYESIAN PREDICTIVE PROBABILITY, SAMPLE SIZE, AND PROTOCOL MODIFICATIONS Considering different degrees of enrollment efficiency and different population
distributions among the treatment arms, we used the Bayesian predictive probability approach to lay out an adaptive design in the new protocol amendment (July 24, 2020). This study was
originally designed to have 10–20 patients enrolled in each arm, bringing the estimated sample size to approximately 140 patients. Referring to the historical data of heavily pretreated TNBC
patients after multiline chemotherapy,42 if three or more patients in each arm reached CR or PR, then the arm would be considered to have reached the efficacy boundary. Using Bayesian
predictive probability, based on the number of patients who achieved objective response (CR + PR) in real time, each arm could be terminated independently according to futility or efficacy
boundaries.43 Assuming that the reference objective response rate is p0 = 15%, the prior probability fits the beta distribution (0.05, 0.05). The final threshold value of 0.5 was adopted for
the arm to achieve effectiveness, 0.1 was adopted as the threshold value for early termination due to ineffectiveness, and 0.9 was adopted as the threshold value for early termination due
to effectiveness. Using Bayesian prediction probability, futility and efficacy boundaries were obtained, and the simulation results under different true values of ORR are shown in the
Supplementary information, Data S1. Due to the difficulty of enrollment and the promising efficacy observed in other arms in the interim analysis, arms A and G could be terminated early at
fewer than 10 patients (more details in Supplementary information, Data S1). Arm C was expanded to a maximum of 41 cases based on the efficacy reported in the interim analysis. Considering a
dropout rate of 10%, 46 patients needed to be enrolled. Some modifications have been made in the new version of protocol, and we briefly summarized them as follows: (1) the sample sizes in
each arm were set to be more flexible according to Bayesian prediction probability method, and the rationale behind it was explained as above, (2) CTCAE version 4.0 was updated to version
5.0, (3) bevacizumab or low dose VEGFR inhibitors have been explored due to the toxicity issues with the VEGFR inhibitors. The study design allowed arms to be dynamic so that old arms could
be eliminated when finished and new arms could be added. Notably, the investigators used FUTURE as a platform to test the safety and efficacy of potential new drug–biomarker combinations in
heavily pretreated TNBC patients. OUTCOMES The primary endpoint was the ORR per investigator according to RECIST v1.1 using imaging at baseline and every two cycles until disease
progression.44 A CR or a PR was confirmed with one sequential tumor assessment at least 4 weeks later. Secondary endpoints were PFS (defined as the interval from the start of treatment to
disease progression or death from any cause, whichever occurred first, or last PFS assessment for patients alive without progression), OS (first study dose until death from any cause),
disease control rate (DCR, proportion of patients who experience a best response of CR or PR or stable disease ≥ 8 weeks according to RECIST version 1.1), and safety and tolerability. For
PFS analysis, death before the first progressive disease (PD) assessment was computed as progressed, death between adequate assessment visits was computed as progressed, and death after more
than one missed visit was censored on the date of last documented nonprogression. Treatment discontinuation for undocumented progression was censored on the date of last assessment without
progression. Safety evaluations included assessments of AEs and serious AEs (SAEs), laboratory safety evaluations, vital signs, and physical examination. AEs were assessed in accordance with
the National Cancer Institute Common Terminology Criteria for AEs, version 5.0. For AEs with various grades, the maximum reported grade was used in the summary table. BIOSPECIMEN
COLLECTION, QUALITY CONTROL, AND PROCESSING Tumor and matched blood DNA were isolated from tumor samples and peripheral lymphocytes using TGuide M24 (Tiangen, Beijing, China). Absorbance at
260 nm (A260) and 280 nm (A280) was measured to estimate the purity and quantity of the total DNA by a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The extracted
DNA was considered suitable for subsequent experiments if the A260/A280 ratio was between 1.6 and 1.9. SEQUENCING USING THE FUSCC-BC PANEL Details on the sequencing protocol have been
described previously.19 The FUSCC breast cancer (FUSCC-BC) panel was used in this study (Supplementary information, Table S7). Both tumor and matched blood samples were sequenced. A KAPA
HyperPlus kit (Kapa Biosystems) and Illumina HiSeq X TEN platform (Illumina Inc., San Diego, CA, USA) were used during NGS sequencing. Each alteration identified by the pipeline was manually
reviewed to ensure that no false positives were reported. GENOMIC BIOMARKER ANALYSIS Somatic mutations were called from the tissue and blood BAM files using GATK4 Mutect2 with the default
parameters. The VCF files were annotated using ANNOVAR. The variants and annotation results were transferred into Excel spreadsheets. Oncogenic signaling pathways were defined based on a
previous study.45 In post hoc exploratory analyses, PFS for each cohort was analyzed by tissue somatic mutation status.19 TNBC ORGANOID AND CELL LINE CLASSIFICATION TNBC organoids were
subjected to IHC staining (AR, CD8 and FOXC1) for subtyping.23 We performed hierarchical clustering to determine the TNBC subtype of common TNBC cell lines based on the similarity of
expression profiles between patients and cell lines (Supplementary information, Fig. S6a). Patient RNA-seq data along with TNBC subtype annotations were obtained from our previous study.18
Cell line RNA-seq data were derived from the Cancer Cell Line Encyclopedia database and the study of Gong and colleagues.46 Cell lines with specific TNBC subtypes (according to the
consistency of the FUSCC TNBC subtype and Lehmann subtype20) were chosen for further experiments. CELL PROLIFERATION ASSAY We used the human TNBC cell lines BT-549, HCC1187, HCC1599, Hs 578
T, MDA-MB-157, MDA-MB-436, and MFM223 (from ATCC). Cell proliferation assays were performed as previously described.47,48 Briefly, the cells of interest (1 × 103–3 × 103 cells per well) were
seeded into 96-well plates overnight in 100 μL of complete growth medium and then treated with the indicated drugs for 5 days in triplicate. Cell viability was tested using the cell
counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Japan, CK04) according to the manufacturer’s instructions. ORGANOID PREPARATION AND CULTURE We developed a biobank for organoid
storage as previously described.46 Briefly, fresh breast cancer tissues were minced into small fragments using sterile scalpels. Tissues were digested and resuspended in 10 mL of TAC buffer,
incubated for 3 min to remove red blood cells and passed through a 100 mm cell strainer (Corning). For passaging, 5 mL of harvesting solution (Trevigen, 3700-100-01) was used to digest the
basement membrane extract, which was incubated on ice for 1 h. Subsequently, the organoids were centrifuged at 350× _g_ for 5 min, washed in digestion buffer and spun down. Next, 3 mL TrypLE
Express (Invitrogen) was added, and organoids were incubated at room temperature for 3 min, followed by mechanical dissociation to small cell clusters by pipetting. Organoids were passaged
at a 1:2–3 dilution every 2–3 weeks. DRUG RESPONSE TEST OF TNBC ORGANOIDS Drug response testing of TNBC organoids was performed according to a previous paper.49 For organoid drug treatment,
organoids in good condition were harvested and digested into single cells. Twenty-five microliters of organoid suspension was added to a cell-repellent black surface in clear bottom 384-well
plates (Greiner 781976-SIN) with 1 × 103–3 × 103 cells per well and cultured for another 5–6 days before drug treatments. Organoids with ADCs were cultured for 2 weeks before testing for
viability. Organoid cell viability was evaluated by a CellTiter-Glo 3D cell viability assay (Promega, G9683) according to the manufacturer’s instructions. PD-L1 AND TROP-2 IHC Baseline PD-L1
expression in the FUTURE trial was assessed at a central laboratory and characterized according to the combined positive score (CPS) as reported previously.11 CPS was defined as the ratio
of PD-L1-positive cells (tumor cells, lymphocytes, and macrophages) out of the total number of tumor cells multiplied by 100. Available tumor specimens were stained for Trop-2 by IHC as
reported previously.37,50 Positivity required at least 10% of the tumor cells to be stained. STATISTICAL ANALYSIS The primary efficacy analysis population was the intention-to-treat
population, including all eligible patients enrolled in the study. Safety was analyzed in all patients who received at least one dose of the study medication. The ORR and DCR with 95% CI
were calculated using the Clopper–Pearson method. PFS and OS with 95% CI were assessed using the Kaplan–Meier method. In post hoc exploratory analyses, PFS for each cohort was analyzed by
tissue somatic mutation. The association between the HER2 IHC scores and TNBC subtypes was examined using the chi-square test and Fisher’s exact test. Two-tailed Student’s _t_-test,
Wilcoxon’s test and Kruskal‒Wallis test were utilized to compare continuous variables where appropriate. All tests were two-sided, and _P_ < 0.05 was regarded as statistically significant
unless otherwise stated. SPSS (version 20) and R (version 4.1.1) were used for statistical analysis. The full statistical analysis plan is available in the protocol. REFERENCES * Waks, A.
G. & Winer, E. P. Breast cancer treatment: a review. _JAMA_ 321, 288–300 (2019). Article CAS PubMed Google Scholar * Global, regional, and national age-sex-specific mortality and
life expectancy, 1950-2017: a systematic analysis for the Global Burden of Disease Study 2017. _Lancet_ 392, 1684–1735 (2018). * Khosravi-Shahi, P., Cabezón-Gutiérrez, L. &
Custodio-Cabello, S. Metastatic triple negative breast cancer: Optimizing treatment options, new and emerging targeted therapies. _Asia Pac. J. Clin. Oncol._ 14, 32–39 (2018). Article
PubMed Google Scholar * Bardia, A. et al. Sacituzumab govitecan in metastatic triple-negative breast cancer. _N. Engl. J. Med._ 384, 1529–1541 (2021). Article CAS PubMed Google Scholar
* Bianchini, G., Balko, J. M., Mayer, I. A., Sanders, M. E. & Gianni, L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. _Nat. Rev. Clin.
Oncol._ 13, 674–690 (2016). Article CAS PubMed PubMed Central Google Scholar * Schmid, P. et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. _N. Engl. J.
Med._ 379, 2108–2121 (2018). Article CAS PubMed Google Scholar * Cortes, J. et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally
recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. _Lancet_ 396, 1817–1828 (2020).
Article PubMed Google Scholar * Liu, J. et al. Multicenter phase II trial of Camrelizumab combined with Apatinib and Eribulin in heavily pretreated patients with advanced triple-negative
breast cancer. _Nat. Commun._ 13, 3011 (2022). Article CAS PubMed PubMed Central Google Scholar * Litton, J. K. et al. Talazoparib versus chemotherapy in patients with germline
BRCA1/2-mutated HER2-negative advanced breast cancer: final overall survival results from the EMBRACA trial. _Ann. Oncol._ 31, 1526–1535 (2020). Article CAS PubMed Google Scholar *
Robson, M. et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. _N. Engl. J. Med._ 377, 523–533 (2017). Article CAS PubMed Google Scholar * Winer, E.
P. et al. Pembrolizumab versus investigator-choice chemotherapy for metastatic triple-negative breast cancer (KEYNOTE-119): a randomised, open-label, phase 3 trial. _Lancet Oncol._ 22,
499–511 (2021). Article CAS PubMed Google Scholar * Ma, D. et al. Molecular features and functional implications of germline variants in triple-negative breast cancer. _J. Natl. Cancer
Inst._ 113, 884–892 (2021). * Modi, S. et al. Trastuzumab deruxtecan in previously treated HER2-low advanced breast cancer. _N. Engl. J. Med._ 387, 9–20 (2022). Article CAS PubMed Google
Scholar * Turner, N. C. et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. _Lancet
Oncol._ 21, 1296–1308 (2020). Article CAS PubMed PubMed Central Google Scholar * Middleton, G. et al. The national lung matrix trial of personalized therapy in lung cancer. _Nature_
583, 807–812 (2020). Article CAS PubMed PubMed Central Google Scholar * Lee, J. et al. Tumor genomic profiling guides patients with metastatic gastric cancer to targeted treatment: The
VIKTORY umbrella trial. _Cancer Discov._ 9, 1388–1405 (2019). Article CAS PubMed Google Scholar * Rothwell, D. G. et al. Utility of ctDNA to support patient selection for early phase
clinical trials: the TARGET study. _Nat. Med._ 25, 738–743 (2019). Article CAS PubMed Google Scholar * Jiang, Y. Z. et al. Genomic and transcriptomic landscape of triple-negative breast
cancers: subtypes and treatment strategies. _Cancer Cell_ 35, 428–440.e5 (2019). Article CAS PubMed Google Scholar * Lang, G. T. et al. Characterization of the genomic landscape and
actionable mutations in Chinese breast cancers by clinical sequencing. _Nat. Commun._ 11, 5679 (2020). Article CAS PubMed PubMed Central Google Scholar * Lehmann, B. D. et al.
Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. _J. Clin. Invest._ 121, 2750–2767 (2011). Article CAS PubMed
PubMed Central Google Scholar * Burstein, M. D. et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. _Clin. Cancer Res._ 21,
1688–1698 (2015). Article CAS PubMed Google Scholar * Wu, S. Y., Wang, H., Shao, Z. M. & Jiang, Y. Z. Triple-negative breast cancer: new treatment strategies in the era of precision
medicine. _Sci. China Life Sci._ 64, 372–388 (2021). Article PubMed Google Scholar * Zhao, S. et al. Molecular subtyping of triple-negative breast cancers by immunohistochemistry:
molecular basis and clinical relevance. _Oncologist_ 25, e1481–e1491 (2020). Article CAS PubMed PubMed Central Google Scholar * Jiang, Y. Z. et al. Molecular subtyping and genomic
profiling expand precision medicine in refractory metastatic triple-negative breast cancer: the FUTURE trial. _Cell Res._ 31, 178–186 (2021). Article CAS PubMed Google Scholar * Berry,
D. A. Adaptive clinical trials in oncology. _Nat. Rev. Clin. Oncol._ 9, 199–207 (2011). Article PubMed Google Scholar * Xu, B. et al. Pyrotinib plus capecitabine versus lapatinib plus
capecitabine for the treatment of HER2-positive metastatic breast cancer (PHOEBE): a multicentre, open-label, randomised, controlled, phase 3 trial. _Lancet Oncol._ 22, 351–360 (2021).
Article CAS PubMed Google Scholar * Tarantino, P. et al. Antibody-drug conjugates: Smart chemotherapy delivery across tumor histologies. _CA Cancer J. Clin._ 72, 165–182 (2022). Article
PubMed Google Scholar * Voorwerk, L. et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. _Nat.
Med._ 25, 920–928 (2019). Article CAS PubMed Google Scholar * Wu, S. Y. et al. Combined angiogenesis and PD-1 inhibition for immunomodulatory TNBC: concept exploration and biomarker
analysis in the FUTURE-C-Plus trial. _Mol. Cancer_ 21, 84 (2022). Article CAS PubMed PubMed Central Google Scholar * Chen, L. et al. Famitinib with camrelizumab and nab-paclitaxel for
advanced immunomodulatory triple-negative breast cancer (FUTURE-C-PLUS): an open-label, single-arm, phase 2 trial. _Clin. Cancer Res._ 28, 2807–2817 (2022). * Hu, X. et al. Multicenter phase
II study of apatinib, a novel VEGFR inhibitor in heavily pretreated patients with metastatic triple-negative breast cancer. _Int. J. Cancer_ 135, 1961–1969 (2014). Article CAS PubMed
Google Scholar * Bose, R. et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. _Cancer Discov._ 3, 224–237 (2013). Article CAS PubMed Google Scholar *
Jhaveri, K. et al. Abstract GS4-10: Neratinib + fulvestrant + trastuzumab for hormone receptor-positive, HER2-mutant metastatic breast cancer and neratinib + trastuzumab for triple-negative
disease: Latest updates from the SUMMIT trial. _Cancer Res._ 82 (4_SUPPLEMENT), GS4-10 (2022). Article Google Scholar * Schmid, P. et al. Capivasertib plus paclitaxel versus placebo plus
paclitaxel as first-line therapy for metastatic triple-negative breast cancer: The PAKT trial. _J. Clin. Oncol._ 38, 423–433 (2020). Article CAS PubMed Google Scholar * Kim, S. B. et al.
Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind,
placebo-controlled, phase 2 trial. _Lancet Oncol._ 18, 1360–1372 (2017). Article CAS PubMed PubMed Central Google Scholar * Bartsch, R. SABCS 2020: update on triple-negative and
metastatic HER2-positive breast cancer. _Memo_ 14, 247–251 (2021). Article PubMed PubMed Central Google Scholar * Bardia, A. et al. Efficacy and safety of anti-Trop-2 antibody drug
conjugate sacituzumab govitecan (IMMU-132) in heavily pretreated patients with metastatic triple-negative breast cancer. _J. Clin. Oncol._ 35, 2141–2148 (2017). Article CAS PubMed PubMed
Central Google Scholar * Bardia, A. et al. Biomarker analyses in the phase III ASCENT study of sacituzumab govitecan versus chemotherapy in patients with metastatic triple-negative breast
cancer. _Ann. Oncol._ 32, 1148–1156 (2021). Article CAS PubMed Google Scholar * Asghar, U. S. et al. Single-cell dynamics determines response to CDK4/6 inhibition in triple-negative
breast cancer. _Clin. Cancer Res._ 23, 5561–5572 (2017). Article CAS PubMed PubMed Central Google Scholar * Gucalp, A. et al. Phase II trial of bicalutamide in patients with androgen
receptor-positive, estrogen receptor-negative metastatic Breast Cancer. _Clin. Cancer Res._ 19, 5505–5512 (2013). Article CAS PubMed PubMed Central Google Scholar * Bonnefoi, H. et al.
A phase II trial of abiraterone acetate plus prednisone in patients with triple-negative androgen receptor positive locally advanced or metastatic breast cancer (UCBG 12-1). _Ann. Oncol._
27, 812–818 (2016). Article CAS PubMed Google Scholar * Cortes, J. et al. Eribulin monotherapy versus treatment of physician’s choice in patients with metastatic breast cancer (EMBRACE):
a phase 3 open-label randomised study. _Lancet_ 377, 914–923 (2011). Article CAS PubMed Google Scholar * Lee, J. J. & Liu, D. D. A predictive probability design for phase II cancer
clinical trials. _Clin. Trials_ 5, 93–106 (2008). Article PubMed PubMed Central Google Scholar * Schwartz, L. H. et al. RECIST 1.1 - standardisation and disease-specific adaptations:
perspectives from the RECIST working group. _Eur. J. Cancer_ 62, 138–145 (2016). Article PubMed PubMed Central Google Scholar * Ding, L. et al. Perspective on oncogenic processes at the
end of the beginning of cancer genomics. _Cell_ 173, 305–320.e10 (2018). Article CAS PubMed PubMed Central Google Scholar * Gong, Y. et al. Metabolic-pathway-based subtyping of
triple-negative breast cancer reveals potential therapeutic targets. _Cell Metab._ 33, 51–64.e9 (2021). * Lewis, J. S. et al. Intrinsic mechanism of estradiol-induced apoptosis in breast
cancer cells resistant to estrogen deprivation. _J. Natl Cancer Inst._ 97, 1746–1759 (2005). Article CAS PubMed Google Scholar * Zhu, X. et al. Efficacy and mechanism of the combination
of PARP and CDK4/6 inhibitors in the treatment of triple-negative breast cancer. _J. Exp. Clin. Cancer Res._ 40, 122 (2021). Article CAS PubMed PubMed Central Google Scholar * Xiao, Y.
et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. _Cell Res._ 32, 477–490 (2022). * Starodub, A. N. et al. First-in-human trial of a novel
anti-Trop-2 antibody-SN-38 conjugate, sacituzumab govitecan, for the treatment of diverse metastatic solid tumors. _Clin. Cancer Res._ 21, 3870–3878 (2015). Article CAS PubMed PubMed
Central Google Scholar Download references ACKNOWLEDGEMENTS The authors would like to extend thanks to all patients, clinicians, and nurses for their efforts in this study. This work was
supported by Jiangsu Hengrui Pharmaceuticals Co. Ltd. (Shanghai, China), grants from the National Key R&D Project of China (2020YFA0112304), the National Natural Science Foundation of
China (91959207 and 92159301), the Shanghai Key Laboratory of Breast Cancer (12DZ2260100), the Shanghai Key Clinical Specialty of Oncology (shslczdzk02001), the Clinical Research Plan of
SHDC (SHDC2020CR4002, SHDC2020CR5005), the Natural Science Foundation of Shanghai (21ZR1414700) and the SHDC Municipal Project for Developing Emerging and Frontier Technology in Shanghai
Hospitals (SHDC12021103). AUTHOR INFORMATION Author notes * These authors contributed equally: Yin Liu, Xiu-Zhi Zhu, Yi Xiao, Song-Yang Wu. AUTHORS AND AFFILIATIONS * Department of Breast
Surgery, Fudan University Shanghai Cancer Center, Shanghai, China Yin Liu, Xiu-Zhi Zhu, Yi Xiao, Song-Yang Wu, Wen-Jia Zuo, Qiang Yu, A-Yong Cao, Jun-Jie Li, Ke-Da Yu, Guang-Yu Liu, Jiong
Wu, Yi-Zhou Jiang, Zhong-Hua Wang & Zhi-Ming Shao * Key Laboratory of Breast Cancer in Shanghai, Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China Yin
Liu, Xiu-Zhi Zhu, Yi Xiao, Song-Yang Wu, Wen-Jia Zuo, Qiang Yu, A-Yong Cao, Jun-Jie Li, Ke-Da Yu, Guang-Yu Liu, Jiong Wu, Yi-Zhou Jiang, Zhong-Hua Wang & Zhi-Ming Shao * Department of
Medical Oncology, Cancer Hospital of China Medical University/Liaoning Cancer Hospital, Shenyang, Liaoning, China Tao Sun * Department of Medical Oncology, First Hospital of Jilin
University, Changchun, Jilin, China Jiu-Wei Cui & Zheng Lv * Department of Breast Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking
University Cancer Hospital & Institute, Beijing, China Hui-Ping Li * Jiangsu Hengrui Pharmaceuticals Co. Ltd, Shanghai, China Xiao-Yu Zhu Authors * Yin Liu View author publications You
can also search for this author inPubMed Google Scholar * Xiu-Zhi Zhu View author publications You can also search for this author inPubMed Google Scholar * Yi Xiao View author publications
You can also search for this author inPubMed Google Scholar * Song-Yang Wu View author publications You can also search for this author inPubMed Google Scholar * Wen-Jia Zuo View author
publications You can also search for this author inPubMed Google Scholar * Qiang Yu View author publications You can also search for this author inPubMed Google Scholar * A-Yong Cao View
author publications You can also search for this author inPubMed Google Scholar * Jun-Jie Li View author publications You can also search for this author inPubMed Google Scholar * Ke-Da Yu
View author publications You can also search for this author inPubMed Google Scholar * Guang-Yu Liu View author publications You can also search for this author inPubMed Google Scholar *
Jiong Wu View author publications You can also search for this author inPubMed Google Scholar * Tao Sun View author publications You can also search for this author inPubMed Google Scholar *
Jiu-Wei Cui View author publications You can also search for this author inPubMed Google Scholar * Zheng Lv View author publications You can also search for this author inPubMed Google
Scholar * Hui-Ping Li View author publications You can also search for this author inPubMed Google Scholar * Xiao-Yu Zhu View author publications You can also search for this author inPubMed
Google Scholar * Yi-Zhou Jiang View author publications You can also search for this author inPubMed Google Scholar * Zhong-Hua Wang View author publications You can also search for this
author inPubMed Google Scholar * Zhi-Ming Shao View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Z.-M.S., Z.-H.W., and Y.-Z.J. conceived and
designed the study. Z.-M.S., Z.-H.W., and Y.-Z.J. supervised the study. Y.L., T.S., J.-W.C., Z.L., H.-P.L., and Z.-H.W. carried out the clinical trial. Y.L., X.-Z.Z. and Q.Y. acquired the
data. Y.L., X.-Z.Z. did the statistical analysis. Y.L. and X.-Z.Z. analyzed and interpreted the data. Y.L., X.-Z.Z., S.-Y.W., Y.X., W.-J.Z., and Y.-Z.J. drafted the manuscript. All authors
critically revised the manuscript. J.-J.L., A.-Y.C., K.-D.Y., G.-Y.L., J.W., T.S., J.-W.C., Z.L., H.-P.L., Z.-M.S., Z.-H.W., and Y.-Z.J. provided resources. Z.-M.S., Z.-H.W., Y.-Z.J., Y.L.
and X.-Y.Z. obtained funding. Z.-M.S., Z.-H.W., and Y.-Z.J. have accessed and verified the data. CORRESPONDING AUTHORS Correspondence to Yi-Zhou Jiang, Zhong-Hua Wang or Zhi-Ming Shao.
ETHICS DECLARATIONS COMPETING INTERESTS X.-Y.Z. is an employee of Jiangsu Hengrui Pharmaceuticals (formerly Jiangsu Hengrui Medicine). All other authors have no conflicts of interest to
declare. SUPPLEMENTARY INFORMATION DATA S1 SUPPLEMENTARY FIGURE 1 SUPPLEMENTARY FIGURE 2 SUPPLEMENTARY FIGURE 3 SUPPLEMENTARY FIGURE 4 SUPPLEMENTARY FIGURE 5 SUPPLEMENTARY FIGURE 6
SUPPLEMENTARY FIGURE 7 SUPPLEMENTARY TABLE 1 SUPPLEMENTARY TABLE 2 SUPPLEMENTARY TABLE 3 SUPPLEMENTARY TABLE 4 SUPPLEMENTARY TABLE 5 SUPPLEMENTARY TABLE 6 SUPPLEMENTARY TABLE 7 RIGHTS AND
PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any
medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The
images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not
included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly
from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Liu, Y., Zhu, XZ.,
Xiao, Y. _et al._ Subtyping-based platform guides precision medicine for heavily pretreated metastatic triple-negative breast cancer: The FUTURE phase II umbrella clinical trial. _Cell Res_
33, 389–402 (2023). https://doi.org/10.1038/s41422-023-00795-2 Download citation * Received: 13 November 2022 * Accepted: 01 March 2023 * Published: 27 March 2023 * Issue Date: May 2023 *
DOI: https://doi.org/10.1038/s41422-023-00795-2 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is
not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
