Peripheral nerve resident macrophages share tissue-specific programming and features of activated microglia
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

Whereas microglia are recognized as fundamental players in central nervous system (CNS) development and function, much less is known about macrophages of the peripheral nervous system (PNS).
Here, by comparing gene expression across neural and conventional tissue-resident macrophages, we identified transcripts that were shared among neural resident macrophages as well as
selectively enriched in PNS macrophages. Remarkably, PNS macrophages constitutively expressed genes previously identified to be upregulated by activated microglia during aging,
neurodegeneration, or loss of Sall1. Several microglial activation-associated and PNS macrophage-enriched genes were also expressed in spinal cord microglia at steady state. We further show
that PNS macrophages rely on IL-34 for maintenance and arise from both embryonic and hematopoietic precursors, while their expression of activation-associated genes did not differ by
ontogeny. Collectively, these data uncover shared and unique features between neural resident macrophages and emphasize the role of nerve environment for shaping PNS macrophage identity.
The significant role of resident neural macrophages in neuroinflammation and disease progression is increasingly appreciated in mouse models and individuals with neurodegeneration1,2,3. Such
advances, which largely rely on the interpretation of data from transcriptional analyses and human genome-wide association studies of Alzheimer’s disease (AD) and other neurodegenerative
conditions, have led to critical findings about cellular and molecular processes underlying such diseases4,5,6. Most of these studies, however, have focused on resident macrophages in the
brain (microglia) and, to a lesser extent, the spinal cord. Meanwhile, the transcriptional identity and functions of resident macrophages in the peripheral nervous system (PNS) remain mostly
unknown.
The PNS consists of a multitude of neuronal networks that relay motor and sensory information between the central nervous system (CNS) and the rest of the body7. Although it has the capacity
to regenerate, the PNS is also prone to injury and degeneration8. Studies of PNS injury have shown that PNS macrophages play important roles for debris clearance, pain development, and
regeneration9,10,11. While the contribution of recruited monocytes cannot be excluded, these studies demonstrate the importance of PNS macrophages in nerve injury. Understanding the roles of
these cells in homeostasis and disease may be broadly beneficial for resolving neuroinflammation.
In addition to monocyte-derived macrophages in nerve injury, there are also resident macrophages in the PNS at steady state12,13. While their residence in neuronal tissues is inherently
microglia-like, PNS macrophages exist within a unique peripheral nerve microenvironment. Moreover, although it is known that CNS microglia are derived from the yolk sac during
embryogenesis14, the ontogeny and developmental requirements of PNS macrophages remains unclear. Considering the growing interest in how tissue environment and ontogeny contribute to
microglial identity and function in neurological diseases, we investigated these questions in PNS macrophages.
Here we show that self-maintaining PNS-resident macrophages are a distinct population with transcriptional signatures relating to specific functions in neuronal tissue. PNS macrophages
significantly express genes that resemble not only homeostatic microglia but also activated microglia from aging and neurodegenerative conditions. We found that PNS macrophages originate
from both embryonic and hematopoietic sources, and exhibit partial dependence on interleukin (IL)-34, an alternative ligand for colony-stimulating factor 1 receptor (CSF1R) that is important
for the development and maintenance of microglia15,16. With the exception of Ccr2 expression, transcriptional signatures in PNS macrophage were largely similar between embryonic and
hematopoietic sources, suggesting that nerve environment controls their identity at steady state.
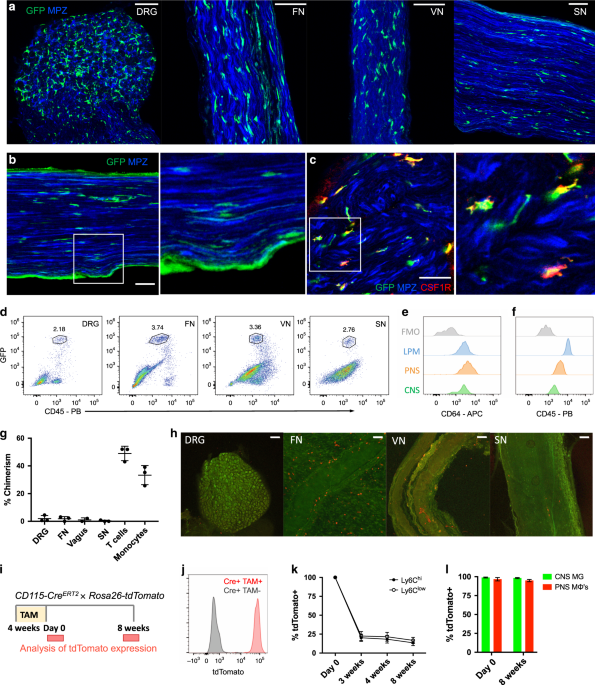
To examine resident macrophages in the PNS, we imaged a variety of nerve types at steady state using CX3CR1GFP/+ reporter mice. In these mice, green fluorescent protein (GFP) effectively
labels microglia and has been shown to label nerve-associated macrophages in adipose, skin, lung, and enteric tissues17,18,19,20,21. CX3CR1GFP/+ cells were found in dorsal root ganglia
(DRG), vagal nerves (VNs), cutaneous intercostal fascial nerves (FNs), and sciatic nerves (SNs) (Fig. 1a). CX3CR1GFP/+ cells were located in the endoneurium (Fig. 1b) and expressed CSF1R,
also known as CD115 (Fig. 1c). Using flow cytometry, we found that CX3CR1GFP/+ cells also expressed the common macrophage marker CD64 (FcγR1)22 and intermediate levels of CD45 (Fig. 1d–f).
Thus, CX3CR1GFP/+ cells in peripheral nerves are indeed macrophages with some resemblance to CNS microglia based on both endoneurial localization and surface marker expression.
a–c Representative confocal imaging of peripheral nerves from CX3CR1GFP/+ MPZtdTomato mice (tomato from MPZ depicted here in blue). a Images of whole-mount dorsal root ganglia (DRG),
cutaneous fascial (FN), vagal (VN), and sciatic nerves (SN) isolated from CX3CR1GFP/+ MPZtdTomato mice. Scale bars are 50 μm. b Endoneurial localization of GFP+ cells in longitudinal
sections of sciatic nerves from CX3CR1GFP/+ MPZtdTomato mice. c CSF1R (red) and CX3CR1GFP (green) colocalization in sciatic nerve cross sections. Scale bars are 50 μm. d Flow cytometric
gating of CX3CR1GFP/+ cells from peripheral nerve tissues. e, f Representative expression of CD64 and CD45 in CX3CR1GFP/+ cells compared with brain microglia, large peritoneal macrophages
(LPMs), and fluorescence minus one (FMO) control. g Flow cytometric quantification of CD45.1 and CD45.2 chimerism in blood (total T cells or total monocytes) and nerves from three pairs of
wild-type (CD45.1) and Lyz2Cre tdTomato (CD45.2) parabionts 10 weeks after joining. h Representative imaging in peripheral nerves from wild-type parabiont; Scale bars are 100 μm. i–l
Analysis of tdTomato expression in tamoxifen-pulsed CSF1RMer-iCre-Mer × Rosa26-tdTomato mice. i Tamoxifen delivery schematic for fate mapping. Mice were given tamoxifen diet for 4 weeks and
analyses for PNS macrophages and CNS microglia were performed at 0 days and 8 weeks after tamoxifen diet removal. j tdTomato expression by genotype from combined peripheral nerves in
tamoxifen-fed mice. k Flow cytometric quantification of Ly6c high and Ly6C low monocytes from mice bled at 0 days, 3 weeks, 4 weeks, and 8 weeks after tamoxifen removal. l Flow cytometric
quantification of CNS microglia (brain and spinal cord) and PNS macrophages (pooled from DRG, fascial nerve, vagus nerve, and sciatic nerve to increase yield in analysis) 0 days and 8 weeks
following tamoxifen removal. Data are mean ± SEM (n = 3 mice per time point). Source data are available as a Source Data file.
To determine whether PNS macrophages depend on circulating precursors or are maintained via local signals, we performed parabiosis in CD45.1+ wild type and CD45.2+ Lyz2Cre × tdTomatofl/fl
mice, and assessed the extent to which cells circulating from the parabiotic partner gave rise to PNS macrophages. Ten weeks after joining the parabionts, we found minimal exchange of PNS
macrophages in all of the nerve types examined, whereas blood T cells and monocytes exchanged robustly (Fig. 1g and Supplementary Fig. 1). Indeed, most of the tdTomato+ cells that could be
seen in the wild-type parabiont were localized to the tissue surrounding the nerves (Fig. 1h). We also performed pulse-chase labeling of PNS macrophages using tamoxifen-inducible
CSF1RMer-iCre-Mer × dTomatofl/fl mice. In these mice, tdTomato expression persists in self-maintaining cells, but not in monocytes, which mostly turn over by 3–4 weeks after tamoxifen
removal23,24. Heterozygous mice were fed tamoxifen diet for 4 weeks and then switched to normal diet (Fig. 1i). Just following tamoxifen removal, 96% of PNS macrophages (pooled from all PNS
sites), 99% of CNS microglia, and 100% of blood monocytes were tdTomato+ (Fig. 1j–l and Supplementary Fig. 2). Whereas only 20% of nonclassical and classical monocytes were still tdTomato+
by 3 weeks after tamoxifen removal, 98% of CNS microglia and 95% of pooled PNS macrophages remained labeled up to 8 weeks following tamoxifen removal (Fig. 1l and Supplementary Fig. 2).
Taken together, these results indicate that PNS macrophages are mostly self-maintained in adult mice.
As we and others have previously demonstrated, unique gene expression profiles can be obtained in tissue-resident macrophage populations across tissue types22,25. To identify signature genes
in peripheral nerve macrophages, we performed bulk RNA sequencing (RNA-seq) to compare purified PNS resident macrophages sorted from DRG, VN, cutaneous intercostal FN, and SNs
(Supplementary Fig. 3) with CNS microglia from the brain and spinal cord, as well as previously characterized conventional macrophage populations from the spleen, peritoneal cavity, and
lungs. Global transcriptomic analysis revealed similarities within resident neural macrophages from both PNS and CNS, with PNS macrophages clustering more closely to CNS microglia than to
conventional macrophages (Fig. 2a and Supplementary Data 1). A substantial number of genes were uniquely enriched in PNS macrophages and CNS microglia compared with the other tissue-resident
macrophages, including microglial signature genes Tmem119, P2ry12, Siglech, Trem2, and Olfml3 (Fig. 2b, c). PNS macrophage-specific genes were also identified (Fig. 2b).
a Sample correlation plot showing global transcriptomic analysis and hierarchical clustering of resident macrophages from PNS, CNS, and conventional macrophages. Each box represents one
replicate. Three replicates comprising up to 20 mice per replicate were included for each population. b Visualization of PNS macrophage unique transcripts (upper quadrant), CNS microglia
unique transcripts (right quadrant), shared transcripts between PNS macrophages and CNS microglia (diagonal, top right quadrant), and conventional macrophages (bottom right quadrant). c
Expression of microglial core transcripts in combined PNS macrophages compared with combined conventional macrophages. Multiple t-tests. Data are mean ± SEM. *P
