Sugar transporter modulates nitrogen-determined tillering and yield formation in rice
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Nitrogen (N) fertilizer application ensures crop production and food security worldwide. N-controlled boosting of shoot branching that is also referred as tillering can improve
planting density for increasing grain yield of cereals. Here, we report that Sugar Transporter Protein 28 (OsSTP28) as a key regulator of N-responsive tillering and yield formation in rice.
N supply inhibits the expression of _OsSTP28_, resulting in glucose accumulation in the apoplast of tiller buds, which in turn suppresses the expression of a transcriptional inhibitor _ORYZA
SATIVA HOMEOBOX 15_ (_OSH15_) via an epigenetic mechanism to activate gibberellin 2-oxidases (GA2oxs)-facilitated gibberellin catabolism in shoot base. Thereby, OsSTP28-OSH15-GA2oxs module
reduces the level of bioactive gibberellin in shoot base upon increased N supply, and consequently promotes tillering and grain yield. Moreover, we identify an elite allele of _OsSTP28_ that
can effectively promote N-responsive tillering and yield formation, thus representing a valuable breeding target of N use efficiency improvement for agricultural sustainability. SIMILAR
CONTENT BEING VIEWED BY OTHERS IMPROVING RICE NITROGEN-USE EFFICIENCY BY MODULATING A NOVEL MONOUNIQUITINATION MACHINERY FOR OPTIMAL ROOT PLASTICITY RESPONSE TO NITROGEN Article 05 October
2023 THE ELITE HAPLOTYPE _OSGATA8_-H COORDINATES NITROGEN UPTAKE AND PRODUCTIVE TILLER FORMATION IN RICE Article Open access 13 June 2024 FERTILIZATION CONTROLS TILLER NUMBERS VIA
TRANSCRIPTIONAL REGULATION OF A _MAX1_-LIKE GENE IN RICE CULTIVATION Article Open access 08 June 2023 INTRODUCTION Nitrogen (N) as an essential macronutrient largely determines crop yield in
agricultural production1. Widespread adoption of high-yielding semi-dwarf gramineous crop varieties after green revolution in 1960s exhibit remarkably enhanced lodging resistance to
elevated N fertilizer input which further drives farmers to supply N fertilizers for boosting yield2,3. However, excessive N fertilizer input causes huge energy cost and environmental risks,
including soil acidification, water eutrophication, and greenhouse gas emission4,5. Thus, the identification of new genetic resources and elite germplasms with high N use efficiency (NUE)
and key genes to enhance NUE becomes an important task for the genetic improvement of grain yield and NUE. In rice production, grain yield is majorly determined by three agronomic traits:
tiller number (also referred as panicle number), grain number per panicle, and grain weight. Due to the defect of gibberellin (GA) biosynthesis pathway in rice semi-dwarf varieties, the
response of plant height is less sensitive to increased N supply, while boosting shoot branching/tillering becomes a more promising approach to improve yield under luxury N input
condition2,3. Indeed, the enhancement of tillering makes a major contribution to the improvement of rice yield and NUE in response to increased N fertilizer input6,7. OsTCP19, encoding a TCP
family transcription factor, plays a vital role in controlling the tillering response to N (TRN) in rice. Intriguingly, the elite allele _OsTCP19-H_ with higher NUE is mostly distributed in
wild rice but it has largely been lost in modern Asian cultivated rice7. However, most modern rice cultivars used after green revolution still exhibit a high sensitivity to boost tillering
in response to increased N supply, indicating the existence of an OsTCP19-independent regulatory pathway to control N-responsive tillering in cultivated rice. In rice, tiller originates from
tiller bud at the axil of leaf, and in turn undergoes the outgrowth process to complete its development8,9. Tillering is a complex trait largely influenced by various environmental cues,
especially by soil N availability6. Increased N supply boosts rice tiller number by promoting the outgrowth rather than the initiation of tiller buds, thus substantially improving effective
panicle number to achieve higher grain yield10,11. External cues of N availability can be integrated into the internal tillering programming via modulating several phytohormone pathways,
such as auxin, strigolactone (SL), and GA6,11,12. In particularly, GA functions as an inhibitor of tiller development13,14. N-mediated Tiller Growth Response 5 (NGR5), a downstream component
of GA signaling pathway, is required for achieving N-induced tillering in rice. NGR5 is a target of GA receptor Gibberellin Insensitive Dwarf 1 (GID1), thus being stabilized by DELLA
proteins. In response to elevated N supply, NGR5 recruits Polycomb Repressive Complex 2 (PRC2) to suppress the expression of several tillering inhibitors via H3K27 methylation, thereby
promoting rice tillering and grain yield11. However, it is still unknown how rice integrates the signal of N availability into the GA-NGR5 module to regulate N-responsive tillering. Here, we
report the identification of a sugar transporter OsSTP28 as a key regulator of N-responsive tillering and yield formation in rice, and demonstrate that N supply represses the expression of
_OsSTP28_ to fine turn the level of apoplastic glucose in tiller buds, which in turn coordinates GA catabolism to boost tillering. In addition, we also identify an elite allele of _OsSTP28_
that can more efficiently promote N-responsive tillering and yield formation in modern cultivated rice varieties, thus providing a valuable germplasm for the genetic improvement of yield and
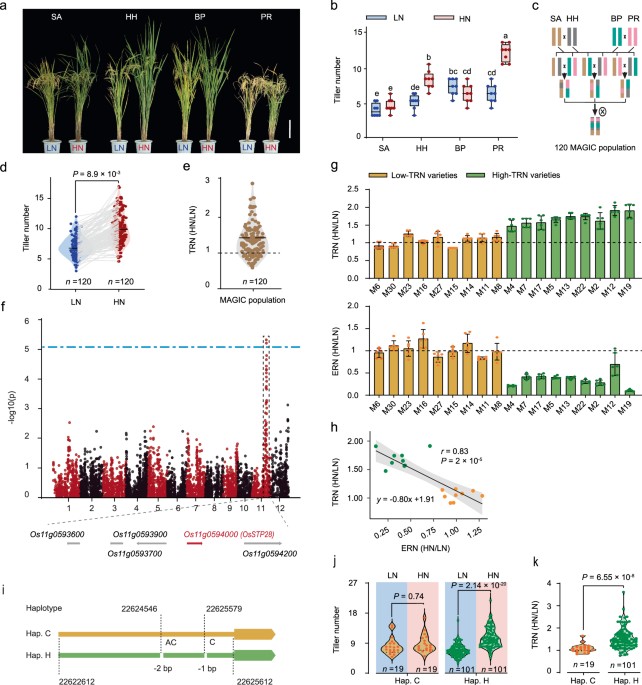
NUE in rice. RESULTS OS_STP28_ IS ASSOCIATED WITH TILLERING RESPONSE TO N IN RICE To explore the genetic basis of TRN, we employed a rice multiparent advanced generation intercross (MAGIC)
population that was developed by four-way cross of four elite rice varieties with a high range of genetic diversity15 (Fig. 1a–c). Firstly, we evaluated the variation of TRN in four parental
varieties in paddy field supplied with low N (LN, 150 kg ha−1) or high N (HN, 350 kg ha−1). Tiller number of two varieties HHZ5-SAL9-Y3-Y1 (HH) and PR33282-B-8-1-1-1-1-1 (PR) were
substantial increased by improved N input, while the tillering response in the other two varieties SAGC-08 (SA) and BP1976B-2-3-7-TB-1-1 (BP) were insensitive to the changed N availabilities
(Fig. 1a, b). The diverse TRN among parental varieties makes it possible to identify the genetic determinant of N-regulated tillering in this MAGIC population. Next, we measured the
tillering trait in this MAGIC panel consisting of 120 lines (Fig. 1c) under both LN and HN conditions, and observed a high degree of phenotypic variation in the tiller number range and
response to N (Fig. 1d, e). By performing a GWAS analysis, we identified a genetic locus at chromosome 11 with a group of single nucleotide polymorphisms (SNPs) highly associated with the
tiller number under HN condition, and the most significantly associated SNP (-log10 (_P_) = 5.20) is located at chromosome 11: 22627320 (Fig. 1f). This linkage disequilibrium (LD) block
spans an approximately 30-kb region containing five candidate genes (Fig. 1f). To ascertain the causal gene for the associated locus, we selected 18 MAGIC lines with contrasting TRN and
assessed the transcriptional expression pattern of all five candidate genes in shoot base under different N supplies. As shown, only _Os11_ _g0594000 (OsSTP28)_ displayed a N-responsive
expression pattern. Specifically, the transcriptional expression level of _OsSTP28_ was remarkably repressed by improved N supply in high TRN lines, by contrast the expression response of
_OsSTP28_ was insensitive to the changed N input in low TRN lines (Fig. 1g). Notably, among all five candidate genes, only _OsSTP28_ exhibited a negative correlation between the expression
response to N (ERN) and TRN under different N supplies (Fig. 1h; Supplementary Fig. 1), suggesting _OsSTP28_ is likely the causal gene leading to the natural variation of TRN in the MAGIC
population. We further dissected the genetic feature of associated locus with TRN by conducting a haplotype analysis of _OsSTP28_ in MAGIC population. It showed that the nucleotide
polymorphism (two InDels are located at chromosome 11: 22624546 and 22625579, respectively) causing the variation of TRN were located in the promoter region of _OsSTP28_ (Fig. 1i), while no
substitution was found in its coding DNA sequence (CDS). We classified the tested MAGIC population into two groups according to the _OsSTP28_ haplotype distribution (Fig. 1i), and MAGIC
lines containing _OsSTP28_ haplotype H (high response to N) allele displayed a higher sensitivity of TRN than those lines with _OsSTP28_ haplotype C (common response to N) allele (Fig. 1j,
k). This result confirmed that _OsSTP28_ is the causal gene determining the genetic diversity of N-induced tillering in rice. To dissect the mechanism why _OsSTP28__Hap. H_ allele is more
sensitive to N supply, we performed a cis-acting element analysis by scanning the promoter regions of _OsSTP28__Hap. C_ and _OsSTP28__Hap. H_. Interestingly, a CCA1/Nhd1 (circadian
regulator) binding element16,17 and a NLP3 (N signaling regulator) binding element18,19 were identified nearby the polymorphic site P1 and P2, respectively (Supplementary Fig. 2a),
indicating these two SNPs in the promoter region of _OsSTP28_ are likely to influence the rhythmic and N-responsive expression pattern of _OsSTP28_. Next, by transient dual-luciferase assays
in rice protoplast, we found that introducing P2Hap. H (Hap.Cp2H) rather than P1Hap. H (Hap.Cp1H) to _OsSTP28__Hap. C_ promoter endowed _OsSTP28_ with a higher sensitivity to N supply
(Supplementary Fig. 2b, c). Moreover, we found that several representative MAGIC lines containing P2Hap. H in _OsSTP28_ promoter exhibited higher sensitivities to N compared with those lines
with P1Hap. H sequence (Supplementary Fig. 2d, e). These results revealed that the polymorphic site P2 with 2-bp indel in _OsSTP28__Hap. H_ promoter is critical to control the
transcriptional response of _OsSTP28_ to N supply in rice. To address the question whether _OsSTP28_ alleles had been selected during rice domestication, we screened _OsSTP28_ haplotype
information among 4726 rice accessions by the algorithm of RiceVarMap v2.020, and found that _OsSTP28__Hap. H_ allele existed in 78.2% of _indica_ rice varieties, 76.6% of _japonica_ rice
varieties, 85.4% of _intermediate_ rice varieties, and 40.4% of _aus_ rice varieties (Supplementary Fig. 3). The frequencies of _OsSTP28__Hap. H_ allele occurrence are nearly equivalent in
_indica_ and _japonica_ rice, implying that _OsSTP28_ might be involved in a conservative regulatory pathway that widely exists across modern Asian cultivated rice and ensures the adaptation
of tillering to the fluctuating N availabilities. _OSSTP28_ ACTS AS A REPRESSOR OF N-DEPENDENT TILLERING IN RICE Next, we assessed the expression pattern of _OsSTP28_ in rice. Quantitative
reverse transcription polymerase chain reaction (qRT-PCR) assay showed that _OsSTP28_ is preferentially expressed in shoot base and slightly expressed in leaf blades of rice seedlings
(Supplementary Fig. 4a). In agreement with this, histochemical analysis of _proOsSTP28:GUS_ reporter lines revealed that _OsSTP28_ is highly expressed in axillary buds with minor expression
in adventitious root primordia and vascular tissue of leaves (Supplementary Fig. 4b). Further, in situ hybridization analysis also confirmed that _OsSTP28_ is preferentially expressed in the
axillary bud (Fig. 2a), indicating a possible function of OsSTP28 in tiller development. Moreover, _OsSTP28_ expression level was induced by N deficiency and repressed by N supply no matter
in the forms of NH4+, NO3−, or NH4NO3 (Fig. 2b), indicating that _OsSTP28_ transcript abundance is determined by N status in seedlings. Indeed, the supply of organic N form glutamine (Gln)
also significantly inhibited the expression of _OsSTP28_ (Fig. 2b). Consistently, when N assimilation was blocked by the supplementation of methionine sulfoximine (MSX), an inhibitor of
glutamine synthetase, the inhibitory effect of N on _OsSTP28_ transcription was largely alleviated (Supplementary Fig. 4c). These results revealed that _OsSTP28_ expression is negatively
regulated by internal N status in rice. Altogether, the tiller bud localization and N-responsive expression pattern strongly suggested an involvement of _OsSTP28_ in the regulation of
N-determined tillering in rice. To explore the function of _OsSTP28_ in tillering, three independent _OsSTP28_ knockout mutants (_stp28-1_, _stp28-2_ and _stp28-3_) were generated in the
background of Nipponbare (NIP) by CRISPR/Cas9 technology (Supplementary Fig. 5a). Knockout of _OsSTP28_ remarkably stimulated the outgrowth of tiller buds (Supplementary Fig. 5b–d).
Consistently, in paddy field, _stp28_ knockout lines also generated more tillers than WT after 6 weeks of transplanting under LN or HN supply (Fig. 2c, d). At the ripening stage (11 weeks
after transplanting), tiller number in _stp28_ mutant lines was increased by ∼49% under LN supply and by ∼35% under HN condition compared with that in WT (Fig. 2c, d), suggesting that
OsSTP28 is a repressor of tillering in rice. Additionally, to evaluate whether TRN in rice depends on the action of _OsSTP28_, we calculated the ratio of tiller number between HN and LN
conditions in WT and _stp28_ mutants. It showed that tillering response in _stp28_ mutants became less sensitive to N supply than that in WT (Fig. 2e). Therefore, our data demonstrate that
OsSTP28 is a negative regulator of tillering and required for achieving tillering response to N in rice. Considering that allelic variation of _OsSTP28_ that occurs in its promoter is the
cause of TRN diversity in rice, we conducted a promoter swapping experiment to verify the differential effect of _OsSTP28__Hap. H_ and _OsSTP28__Hap. C_ alleles on N-dependent tillering. For
this purpose, we complemented _stp28-3_ mutant with _OsSTP28_ coding sequence (cloned from NIP variety) driven by the promoter of _OsSTP28__Hap. H_ or _OsSTP28__Hap. C_ allele (Fig. 2f).
The complementary construct of _OsSTP28_ alleles recovered the expression of _OsSTP28_ in _stp28_ mutant, but interestingly _OsSTP28__Hap. H_ allele was responsive to N supply at
transcriptional level whereas _OsSTP28__Hap. C_ was nearly insensitive to N under the same condition (Fig. 2h). Accordingly, the complementation of _OsSTP28_ alleles recovered tiller
development in _stp28_ mutant, but _OsSTP28__Hap. H_ rather than _OsSTP28__Hap. C_ allele endowed _stp28_ mutant with the ability to achieve N-induced tillering to the similar extent as
wild-type (Fig. 2i). The significantly negative correlation between _OsSTP28_ transcript abundance and tiller number among all these lines highly suggested that the expression level of
_OsSTP28_ determines TRN (Fig. 2j), and in addition, _OsSTP28__Hap. H_ allele exhibited a higher sensitivity of TRN than that of _OsSTP28__Hap. C_ allele (Fig. 2k). Thereby, we conclude that
_OsSTP28__Hap. H_ is an elite allele to regulate TRN in rice. _OSSTP28_ ENCODES AN INFLUX HEXOSE TRANSPORTER COORDINATING N-MEDIATED RHYTHMIC FLUCTUATION OF GLUCOSE IN RICE Sugar
Transporter Protein (STP) is a prominent member of monosaccharide transporter family in plants21. Here, OsSTP28 is predicated as a STP member in rice (Supplementary Fig. 6a). To verify the
function of OsSTP28 as a hexose transporter, we assessed its subcellular localization in rice protoplast and its activity in transporting different forms of hexose in yeast cells. The
results show that OsSTP28 is a plasma membrane localized protein (Fig. 3a) and exhibits higher capacity to transport glucose, lower capacity to transport mannose and galactose, and no
activity in transport of fructose (Supplementary Fig. 7a). 13C-glucose uptake experiment conducted in yeast (Fig. 3b) and in _Xenopus laevis_ oocyte (Fig. 3c, d) further revealed that
OsSTP28 mediates glucose influx rather than efflux. Moreover, kinetic analysis of 13C-glucose showed that OsSTP28 facilitates high affinity glucose uptake with a _K__m_ = 76.15 ± 5.54 µM
(Supplementary Fig. 7b). Taken together, our data demonstrate that OsSTP28 is a plasma membrane-localized hexose transporter responsible for glucose influx. Next, we addressed the question
whether OsSTP28 impacts sugar status or distribution in the shoot base where the tiller is initiated. Interestingly, _OsSTP28_ expression in the shoot base displayed a strong rhythmic
pattern, which peaked in the night and troughed in the day (Fig. 3e). Under the same condition, sugars in WT and _stp28_ mutants also exhibited the rhythmic fluctuations with sucrose peaked
at dusk and troughed at dawn. Notably, glucose peaked at middle of daytime, troughed at the onset of night phase. Both glucose and fructose increased during night (Fig. 3f). Particularly,
rather than sucrose and fructose, the status of glucose was most significantly influenced by the knockout of _OsSTP28_ representing as an enhancement of glucose over the night period without
altering the phase of its diel oscillation (Fig. 3f). These data revealed a vital role of OsSTP28 in modulating glucose status of the shoot base at night. Then, we detected the effect of
OsSTP28 on N-mediated sugar distribution in the shoot base. The level of glucose and fructose in the shoot base was elevated in the night and reduced in the day, while sucrose exhibited an
opposite trend (Supplementary Fig. 8a), supporting the common observation that sucrose is enzymatically hydrolyzed to glucose and fructose for the respiration to maintain plant growth in the
evening22. Elevated N supply increased both sucrose and glucose of WT at day and/or night. Compared with WT, knockout of _OsSTP28_ led to a remarkable accumulation of glucose, but not
sucrose or fructose, in the shoot base at night irrespective of N supply level (Supplementary Fig. 8a). 13C-glucose feeding experiment further revealed that the blade-to-shoot base glucose
translocation was enhanced by N supply in WT, and significantly improved by the knockout of _OsSTP28_ (Supplementary Fig. 8b). Since _OsSTP28_ expression was repressed by N supply (Fig. 2b),
we assumed that N-induced glucose accumulation in shoot base was likely due to the down-regulation of _OsSTP28_, and thereby _OsSTP28_ inactivation further elevated the accumulation of
glucose in the shoot base. This hypothesis was verified by the experiment that complementation of _OsSTP28_ expression in _stp28_ mutant recovered the concentration of both endogenous and
external 13C-labelled glucose in the shoot base to the same extent as WT, especially driven by the promoter of elite _OsSTP28__Hap. H_ allele (Supplementary Fig. 8c, d). We further addressed
the question how OsSTP28 impacts sugar distribution associated with tillering development. Given that OsSTP28 facilitates glucose influx on plasma membrane, we hypothesized that
down-regulation or knockout of _OsSTP28_ would lead to an accumulation of glucose in the apoplast. Indeed, apoplastic glucose level in shoot base was increased by HN supply in WT, and this
accumulation of apoplastic glucose was further dramatically enhanced by knockout of _OsSTP28_ irrespective of N conditions (Fig. 3h). Notably, the change of apoplastic glucose between LN and
HN supplies or between WT and _stp28_ mutants was observed only at night not at the daytime. Meanwhile, no significant difference of apoplastic sucrose or fructose was observed between WT
and _stp28_ mutants (Fig. 3g, h). By contrast, intracellular sugar concentration, no matter for sucrose, glucose or fructose, did not show significant difference between WT and _stp28_
mutants (Supplementary Fig. 9). Altogether, our study demonstrates that OsSTP28 plays a critical role in regulating the N-responsive rhythmic change of apoplastic glucose in rice shoot base.
Since _OsSTP28_ is also slightly expressed in leaf blade, we further monitored apoplastic sugar status in leaves. The change of apoplastic sugar in leaf blade exhibited the same trend as
that in shoot base. Specifically, at night, apoplastic glucose in leaf blade was also enhanced by N supply in WT, and the level of apoplastic glucose was elevated in _stp28_ mutants compared
with that in WT irrespective of N condition (Supplementary Fig. 10). Since N supply or knockout of _OsSTP28_ improved blade-to-shoot base glucose translocation (Supplementary Fig. 8b),
OsSTP28-depedent accumulation of apoplastic glucose in leaves might contribute to establish the glucose pool nearby axillary buds which is critical for stimulating tillering in rice. OSSTP28
COORDINATES GIBBERELLIN METABOLISM TO REGULATE TILLERING RESPONSE TO N To uncover the molecular mechanism underlying OsSTP28-regulated TRN in rice, we performed RNA-sequencing (RNA-seq)
analysis of shoot base between WT and _stp28_ mutant under LN and HN supplies. A large number of differentially expressed genes (DEGs) between WT and _stp28_ mutant (LN condition: 2466 DEGs;
HN condition: 1744 DEGs) were identified (Supplementary Fig. 11a, b; Supplementary Data 1). Among them, many previously characterized tillering regulatory genes were differentially
expressed between WT and _stp28_ mutant and affected by N supplies. For instance, positive tillering regulators (_MOC1_, _OsMASD57_) were significantly up-regulated in _stp28_ mutant, while
negative tillering regulators (_OsTB1_, _OsSPL14_) were down-regulated in _stp28_ mutant in comparison to WT23,24,25,26 (Supplementary Fig. 12–13; Supplementary Data 2). The expression
patterns of these tillering regulators were in well agreement with the enhanced tillering phenotype in _stp28_ mutant compared with WT under both LN and HN supplies (Fig. 2c, d). Gene
ontology (GO) enrichment analysis of DEGs (_stp28_ mutant _vs_ WT) under LN and HN conditions further revealed that many biological processes relating to carbohydrate metabolism, glucose
metabolism, carbon fixation, and organ growth were affected by the knockout of _OsSTP28_ (Supplementary Fig. 11c; Supplementary Data 3), consistent with the elevated apoplastic glucose
accumulated in the shoot base of _stp28_ mutant (Fig. 3h). Remarkably, GO enrichment analysis indicated that several GA metabolic processes were enriched among the DEGs (_stp28_ mutant _vs_
WT) under different N supplies (Supplementary Fig. 11c). Gene set enrichment analysis (GSEA) further highlighted that GA catabolism process was activated by the knockout of _OsSTP28_ in rice
under both LN and HN conditions (Supplementary Fig. 11d; Supplementary Data 4). Accordingly, the transcript abundances of a group of _GA2-oxidases_ (_OsGA2ox3_, _OsGA2ox5_, _OsGA2ox8_ and
_OsGA2ox9_) that catalyze GA catabolism in rice were remarkably increased in _stp28_ mutant compared with WT irrespective of N supply conditions (Fig. 4a; Supplementary Fig. 11e, f). In
agreement with the expression pattern of _GA2-oxidases_, the GA2-oxidase enzymatic activity was increased in _stp28_ mutants compared with WT (Fig. 4b). Since GA2-oxidase catalyzes the
deactivation of bioactive GAs (GA1 and GA4) to generate catabolic GAs (GA8 and GA34) in plants27, we assumed that the activation of GA2-oxidases in _stp28_ mutants might change GA status.
Indeed, bioactive GA1 level was reduced in the shoot base of _stp28_ mutants compared with WT, while catabolic GA8 level was increased at the same situations (Fig. 4d). Besides, bioactive
GA4 and its catabolic form GA34 were too litter to be detected in our conditions (Supplementary Fig. 14). Meanwhile, in WT rice, increased N supply also enhanced the expression of
_OsGA2ox3/5/8_ and the enzymatic activity of GA2-oxidases, thus leading to a reduction of bioactive GA level in the shoot base (Fig. 4a–d). These results demonstrate that increasing N supply
or knockout of _OsSTP28_ reduces bioactive GA level via stimulating GA catabolism in rice, suggesting a vital role of OsSTP28 in the modulation of N-responsive GA metabolism. Since GA acts
as a repressor of N-responsive tillering in rice11, we further investigated the relationship between OsSTP28 and GA metabolism in tillering regulation. _OsGA2ox5_ had a highest transcript
abundance among four OsSTP28-regulated GA2-oxidases (Fig. 4a; Supplementary Fig. 11f). Thus, we generated the knockout mutants of _OsGA2ox5_ (_ga2ox5-1, ga2ox5-2, and ga2ox5-3_) by
CRISPR/Cas9 system (Supplementary Fig. 15a). As expected, knockout of _OsGA2ox5_ substantially reduced GA2-oxidases enzyme activity in shoot base (Fig. 4f), which was supposed to cause an
over-accumulation of bioactive GA in seedlings. Correspondingly, tiller number and TRN were repressed by knockout of _OsGA2ox5_ in rice (Fig. 4e–g). More importantly, we found that the
enhanced tillering phenotype of _stp28_ mutant was completely abolished in _stp28/ga2ox5_ double mutant, suggesting that OsSTP28 functions genetically upstream of GA2-oxidase to regulate
N-responsive tillering. Since NGR5 is a downstream component of GA pathway, NGR5 promotes N-responsive tillering by suppressing the expression of several tillering inhibitory genes, e.g.
OsSPL14, D1411. By generating two _ngr5_ mutants and a double mutant of _stp28/ngr5_ in NIP background, we found that _stp28/ngr5_ displayed a highly inhibited N-responsive tillering
phenotype nearly the same as that of _ngr5_ single mutant rather than _stp28_ mutant (Supplementary Fig. 16), indicating that OsSTP28 functions genetically upstream of NGR5 as well. Taken
together, our study demonstrates that OsSTP28 mediates a regulatory event that functions upstream of GA metabolism and NGR5 pathway to modulate N-dependent tillering in rice. OSH15 IS
CRITICAL FOR OSSTP28-REGULATED GIBBERELLIN CATABOLISM AND TILLERING RESPONSE TO N Since OsSTP28-regulated GA catabolism was achieved by altering _OsGA2oxs_ expression at transcriptional
level, we supposed that a transcription factor is required for this process. By data mining in the RNA-seq results, a _knotted1_-like homeobox (KNOX) transcription factor, _Oryza sativa
homeobox 15_ (_OSH15_) was identified as the most sensitive DEG between _stp28_ mutant and WT under both LN and HN conditions (Fig. 5a). Specifically, _OSH15_ expression was dramatically
declined in _stp28_ mutant compared with WT (Fig. 5b). Since the promoter of most GA2-oxidase genes including _OsGA2ox3, OsGA2ox5, OsGA2ox8, and OsGA2ox9_ present TGAC motifs, a conserved
KNOX binding site (Fig. 5c), OSH15 might be directly involved in the OsSTP28-regulated GA catabolism. Electrophoretic mobility shift assay (EMSA) revealed that OSH15 directly bound to the
promoters of _OsGA2ox3, OsGA2ox5, OsGA2ox8, and OsGA2ox9_ (Fig. 5c), thus resulting in a strong inhibition effect of OSH15 on the expression of _OsGA2ox3, OsGA2ox5, OsGA2ox8, and OsGA2ox9_
(Fig. 5d). To further investigate the function of OSH15 in GA metabolism and tillering, we generated _osh15_ mutant lines by CRISPR/Cas9 system (Supplementary Fig. 15b). As expected, the
expression of _OsGA2ox3, OsGA2ox5, OsGA2ox8, and OsGA2ox9_ as well as the activity of GA2-oxidase were largely enhanced by the knockout of _OSH15_ (Fig. 5e, f), and correspondingly tillering
was also remarkably stimulated in the _osh15_ mutants (Fig. 5g, h), fully mimicking the phenotype of _stp28_ mutants. These results suggested that OSH15 makes a major contribution to
achieve the influence of OsSTP28 on GA catabolism and tillering via inhibiting the transcription of _OsGA2oxs_. In agreement with this point, the complementation of _OsSTP28_ in _stp28_
mutant, especially _OsSTP28__Hap. H_ allele, recovered the expression of _OSH15_ and _OsGA2oxs_ as well as the activity of GA2-oxidase to the same extent as WT (Supplementary Fig. 17a, c),
confirming that OSH15 functions downstream of OsSTP28 to regulate GA catabolism and tillering in rice. Moreover, _OSH15_ expression is inhibited by elevated N supply in WT rice (Fig. 5b),
and the responses of GA catabolism and tillering to N supply in _osh15_ mutants were insensitive to the changed N inputs compared with WT (Fig. 5e–h), revealing a vital role of OSH15 in
modulating N-responsive tillering in rice. To understand the molecular mechanism of OsSTP28-regulated TRN in rice, it is important to address the question how OSH15 is repressed by N supply
or by knockout of _OsSTP28_. Given that OsSTP28 largely determines the glucose level in tiller buds, we thought that the repression of _OSH15_ under HN supply or in _stp28_ mutant might be
relate to the effect of glucose signal. Indeed, we found that many components of glucose signaling pathway such as _OsHKXs_ and _OsTOR_ were remarkably activated upon HN supply or in _stp28_
mutant, while carbon deficiency regulators _OsSnRKs_ were obviously suppressed at the same conditions, indicating that glucose signal is activated by N supply or by knockout of _OsSTP28_ in
rice (Supplementary Fig. 18a, b). Recently, it has been reported that glucose signal causes epigenetic silencing of many key genes by activating PRC2-facilitated trimethylation of histone
H3 at Lysine 27 (H3K27me3) in plants28. Interestingly, _KNOX_ (an orthologue of _OSH15_ in Arabidopsis) is also found to be silenced by H3K27me3 modification29. Thus, it is possible that
glucose-dependent epigenetic silencing might inhibit _OSH15_ expression during the response of tillering to N. By remining the epigenome data in rice30 via ChIP-Hub algorithm31, we found a
strong H3K27me3 signal at the coding sequence of _OSH15_, especially within its first three exons (Supplementary Fig. 18c), implying that _OSH15_ has been regulated by epigenetic silencing
in rice. Then, we conducted a H3K27me3-specific chromatin immunoprecipitation PCR (ChIP-PCR) assay and found that the H3K27me3 modification of _OSH15_ was significantly enhanced by HN supply
in WT or by the knockout of _OsSTP28_ irrespective of N condition (Supplementary Fig. 18d), which fully explained the silencing of _OSH15_ at the same conditions (Fig. 5b). Altogether, we
propose that H3K27me3-related epigenetic modification silences _OSH15_ expression during OsSTP28-mediated TRN. Next, we verified whether the elevated apoplastic glucose in _stp28_ mutants or
under HN supply intermediates the impact of OsSTP28 on _OSH15-OsGA2oxs_ module to regulate tillering. For this purpose, we supplied 2% glucose to WT rice in order to mimic the response of
apoplastic glucose to N input or _OsSTP28_ knockout. It showed that the external glucose supplementation significantly stimulated tillering in rice, and consistently _OSH15_ expression was
repressed while _OsGA2oxs_ expression and GA2-oxidase activity were enhanced at the same situation (Supplementary Fig. 19). In addition, 2-Deoxy-d-Glucose (2-DG), a competitive glycolytic
inhibitor, was supplied together with glucose to WT rice. Interestingly, the supply of 2-DG did not weaken but even slightly enhanced the effect of glucose on _OSH15_ expression,
_OsGA2oxs_-mediated GA catabolism, and tillering (Supplementary Fig. 19), suggesting that glucose itself rather than glycolysis acts as a signal to regulate _OSH15-OsGA2oxs_ module thus
impacting tillering in rice. ELITE ALLELE OF _OSSTP28_ CONTRIBUTES TO IMPROVEMENT OF N-RESPONSIVE YIELD FORMATION AND N USE EFFICIENCY IN RICE We carried out field experiment to investigate
the contribution of OsSTP28 in the improvement of yield and NUE in rice production. Among the three determinants of grain yield, the panicle number as a direct reflection of tillering was
largely improved in _stp28_ mutants compared with WT under both LN and HN supplies (Supplementary Fig. 20a, b; Fig. 6a, b). Meanwhile, the grain number per panicle or grain weight was not
increased even reduced at the limited N condition in _stp28_ mutants (Supplementary Fig. 20c, d; Fig. 6c, d), which might reflect a tradeoff of the boosting tillering in _stp28_ mutants. As
a consequence, due to the largely enhanced tillering (panicle number), inactivation of _OsSTP28_ resulted in increase of grain yield and NUE by about 20–21% under LN condition and by about
10-13% under HN condition (Supplementary Fig. 20e, f; Fig. 6e, f). The results confirm that tillering enhancement makes a major contribution to the improvement of rice yield and NUE in
response to increased N fertilizer input2,3,7, and demonstrate that OsSTP28 plays critical role in regulating TRN and yield formation, especially when N application is limited. Moreover, the
complementation of _OsSTP28__Hap. H_ allele achieved a higher panicle number, grain yield, and NUE compared with that of _OsSTP28__Hap. C_ allele under HN supply (Fig. 6a, b, e, f),
demonstrating that the elite _OsSTP28__Hap. H_ allele is a valuable germplasm for the genetic improvement of N-responsive yield formation and NUE in rice. Furthermore, we address the
question whether the modulation of TRN in rice by OsSTP28 is independent of OsTCP19. First, we found that the allelic variation between _OsTCP19-H_ and _OsTCP19-L_ did not alter TRN in our
four-parent MAGIC population (Supplementary Fig. 21a–c). Since the negative transcriptional response of _OsTCP19_ to N supply endows OsTCP19 with the ability to module N-induced tillering7,
we monitored _OsTCP19_ expression pattern in _stp28_ mutants. As shown, N-inhibited expression pattern of _OsTCP19_ was unaffected by the knockout of _OsSTP28_ (Supplementary Fig. 21d), and
N-repressed expression of OsSTP28 was unchanged in the _ostcp19_ mutant compared with its WT either (Supplementary Fig. 21e). These data strongly suggested that OsSTP28 and OsTCP19 works
independently with each other in rice. Interestingly, the elite allele _OsTCP19-H_ has almost been lost in modern Asian cultivated rice varieties, especially in those accessions developed in
N-rich region7. By contrast, the elite _OsSTP28__Hap. H_ allele was highly distributed in Asian cultivated rice (Supplementary Fig. 3), thus implying a possible complementary occurrence of
the _OsSTP28__Hap. H_ allele to that of _OsTCP19-H_. To test this hypothesis, we re-sequenced _OsTCP19_ and _OsSTP28_ alleles in 18 modern elite japonica rice varieties that are currently
used for rice production in Jiangsu Province (a temperate and N-rich region in China). Indeed, all 18 modern elite japonica rice varieties contain the weak allele _OsTCP19-L_, while 17 of
them contains the elite _OsSTP28__Hap. H_ allele (Supplementary Fig. 21f). Among these 18 modern elite japonica rice varieties, the tillering and yield response was still quite sensitive to
increased N fertilizer input (Supplementary Fig. 21g). Thus, the elite allele _OsSTP28__Hap. H_ is likely to complement the declination of _OsTCP19_ to modulate N-responsive tillering and
yield formation in modern elite japonica rice varieties mainly developed and cultivated in N-rich region. In addition, the expression response of _OsSTP28_ to N was negatively correlated
with TRN in these tested rice varieties (Supplementary Fig. 21i), indicating a potential breeding strategy to improve N-responsive tillering and yield via suppressing the transcript
abundance of _OsSTP28_. Indeed, knockout of _OsSTP28_ in several elite Asian cultivated rice such as MH63 (Ming Hui 63, containing _OsSTP28__Hap. C_ allele) and GLA4 (Guang Lu Ai 4,
containing _OsSTP28__Hap. H_ allele) by genome editing substantially improved their N-responsive tillering and grain yield (Fig. 6g, h; Supplementary Fig. 22). Taken together, our study
reveals that _OsSTP28_ is a vital genetic locus with large potential to improve rice yield and NUE in agricultural production by coordinating phytohormone metabolism with N fertilizer input.
DISCUSSION The genetic basis and molecular mechanism of tillering response to N and yield formation are inherently complex. It has been shown that a transcription factor OsTCP19 contributes
to geographical adaptation of rice tillering response to diverse soil N content7. The elite allele _OsTCP19-H_ is enriched in rice varieties originating from N-deficient regions (such as
wild rice or _aus_ rice varieties) and has been lost in most modern cultivated rice varieties. However, modern elite rice cultivars used in current rice production still exhibits high
sensitivity to achieve N-induced tillering (Supplementary Fig. 21g), implying that a OsTCP19-independent pathway is required to modulate tillering response to N. Indeed, our study shows that
OsSTP28 functions independent of OsTCP19 (Supplementary Fig. 21a–e). More importantly, the elite allele _OsSTP28__Hap. H_ displays a higher frequency in modern cultivated rice, thus showing
a complementary occurrence compared with _OsTCP19-H_ (Supplementary Fig. 3). These data suggest that OsSTP28 facilities a OsTCP19-independent regulatory cascade to complement the
declination of _OsTCP19_ and module N-responsive tillering in modern elite rice varieties which are mainly selected and cultivated in N-rich regions (Supplementary Fig. 21f, h). Moreover,
the negative correlation between the expression response of _OsSTP28_ and the tillering response to N in current elite rice cultivars used in China suggests that reducing the expression of
_OsSTP28_ may represent a practical breeding strategy for rice yield and NUE improvement. Green revolution in rice originates from the discovery and application of serious allelic mutations
of _semi-dwarf1 (sd1)_ that causes defects on GA biosynthesis and reduces the abundance of bioactive GA32,33. Thus, GA pathway is tightly linked with rice productive and NUE in particularly
after green revolution2,6. Previous study demonstrates that GA signaling pathway and its downstream component NGR5 play a critical role in achieving N-responsive tillering in rice11. Upon
elevated N availability, NGR5 is stabilized by DELLA proteins to trigger rice tillering. However, it remains unclear how the signal of N availability is integrated into GA-NGR5 pathway to
regulate tiller development. In present study, we find that OsSTP28 acts as a critical intermediator to transduce the signal of N availability to coordinate GA catabolism with tillering
response to N in rice. In response to increased N supply, suppressed expression of _OsSTP28_ leads to the activation of GA2oxs-facilitated GA catabolism in shoot base, which reduces the
content of bioactive GAs and consequently promotes tillering (Fig. 4). This impact of OsSTP28 on GA catabolism depends on the involvement of a transcription inhibitor OSH15 that can directly
bind to the promoter of _GA2oxs_ to negatively regulate GA catabolism (Fig. 5). Intriguingly, OSH15 binding site, TGAC motif, is widely existed in the promoters of _GA2oxs_ across
gramineous species, and similarly, KNOTTED1, an orthologue of OSH15 in maize, is also able to directly repress _GA2oxs_ expression34, suggesting that the regulation of OSH15 on GA catabolism
is likely a conserved mechanism across gramineous species. Furthermore, genetic analysis verified that OsSTP28 functions upstream of GA catabolism and NGR5 to modulate N-responsive
tillering (Fig. 4h–j; Supplementary Fig. 16e, f). Taken together, OsSTP28-OSH15-GA2oxs represents an important entry point of the environmental N availability to coordinate GA-NGR5 pathway
with tiller development in rice. Sugar is an important regulator of shoot branching/tillering across many plant species35. Previous studies elucidate that elevated sugar content in shoot
base where tiller buds are generated can promote tiller outgrowth in rice16,36. In agreement with this point, we observed an accumulation of glucose in the shoot base of rice under high N
supply (Fig. 3g, h; Supplementary Fig. 8), and the supplementation of external glucose stimulate tiller bud outgrowth (Supplementary Fig. 19a, b). Note that, sugar homeostasis in plants is
tightly associated with photoperiod. Carbon dioxide is fixed by photosynthesis to generate sucrose and starch in the light period; whereas starch and sucrose are degraded into glucose and
fructose to maintain respiration in the dark period37. In particular, apoplast sucrose is hydrolyzed by cell wall invertase to produce glucose then imported into sink tissues by STPs38. This
explains why the N-responsive expression of _OsSTP28_ and accumulation of glucose are taken place at night (Fig. 3; Supplementary Fig. 8). Considering that OsSTP28 facilitates glucose
influx, the inactivation of _OsSTP28_ either by increasing N supply or by knockout of _OsSTP28_ leads to the accumulation of apoplastic glucose which functions as a signal to stimulating
tillering in rice (Fig. 3h). Meanwhile, we have not detected significant difference of intracellular glucose between HN and LN or between WT and _stp28_ mutant (Supplementary Fig. 9).
However, it is still a big challenge to precisely detect glucose level in the cytoplasm. Recently, OsSTP15, an orthologue of OsSTP28, has also been reported as a regulator of tillering in
rice36. Compared with _OsSTP28_, we found _OsSTP15_ was insensitive to N supply (Supplementary Fig. 6b). OsSTP15 modules tillering by impacting cytokinin (CK)36, seems not by impacting GA
pathway. These differences between OsSTP15 and OsSTP28 suggest that OsSTP15 is more likely to be a general regulator of tillering, while OsSTP28 is a vital player of N-responsive tillering.
Nevertheless, our study highlights that OsSTP28 serves as a critical checkpoint to coordinate an appropriate balance between N availability and carbohydrate distribution that is essential
for achieving N-responsive tillering and yield formation in rice. Since the influence of glucose on OSH15-GA2oxs mediated GA catabolism and tillering does not rely on the glucose metabolic
process (Supplementary Fig. 19), we consider that glucose acts likely as a signal to coordinate GA catabolism with N-responsive tillering. To evaluate the influence of glucose on
OsSTP28-mediated TRN, we further dissected the expression of several key components in glucose signaling pathway by transcriptome analysis. It is clearly shown that the glucose signal is
activated by increasing N input or by knockout of _OsSTP28_ in shoot base (Supplementary Fig. 18a), which is consistent with the enhanced tillering at the same conditions (Fig. 2c–e;
Supplementary Fig. 5). Although the mechanism in sensing apoplastic glucose level needs to be uncovered, the glucose signal in plant can activate PRC2 complex in a TOR kinase-dependent
manner to promoter H3K27 methylation which suppresses many gene expression28. Our data demonstrate that N-induced and OsSTP28-related silencing of _OSH15_ in rice is due to the activated
H3K27me3 modification (Supplementary Fig. 18c, d). Interestingly, _KNOX_ an orthologue of _OSH15_ in Arabidopsis also undergo a H3K27me3-related epigenetic silencing29. It seems that
H3K27me3 modification functions as a conserved epigenetic mechanism to modulate the expression of _KNOX-like_ transcription factor across the plant kingdom. In summary, our study
demonstrates that OsSTP28 functions as a key regulator to achieve N-responsive tillering and yield formation in rice (Fig. 7). External N input negative regulates the expression of _OsSTP28_
to cause an accumulation of glucose in tiller buds. Then, the elevated glucose signal silences _OSH15_ via H3K27me3 modification to release the expression of GA2oxs that coordinates GA
catabolism to boost tillering. Moreover, an elite allele of _OsSTP28_ with higher sensitivity to N supply or inactivation of _OsSTP28_ are able to promote N-responsive tillering and yield
formation effectively in modern cultivated rice varieties, thus providing a gene resource and the strategy to the genetic improvement of yield and NUE for the sustainable rice production.
METHODS PLANT MATERIALS 120 lines of MAGIC population were developed via four-way cross by using HHZ5-SAL9-Y3-Y1 (HH), PR33282-B-8-1-1-1-1-1 (PR), SAGC-08 (SA) and BP1976B-2-3-7-TB-1-1 (BP)
as the parent varieties15. _Oryza sativa_ (ssp. Japonica. cv. Nipponbare) was used as wild-type rice in this study. _stp28_ mutants (_stp28-1_, _stp28-2_ and _stp28-3_), _osh15_ mutants
(_osh15-1_ and _osh15-2_) and _ga2ox5_ mutants (_ga2ox5-1_, _ga2ox5-2_ and _ga2ox5-3_) were generated in the Nipponbare (NIP) background by CRISPR/Cas9 technology. To generate _OsSTP28_
complementation lines, the coding sequence of _OsSTP28_ from NIP under the control of different promoters (2600 bp-_OsSTP28__Hap. C_ from SA; 2600 bp-_OsSTP28__Hap. H_ from HH) was
introduced into _stp28-3_ mutant via infection with A. tumefaciens strain EHA105. To knockout _OsSTP28_ in the elite cultivated rice varieties, _stp28__GLA4_ and _stp28__MH63_ mutants were
generated in the background of GLA4 (Guang Lu Ai 4, _indica_ rice) and MH63 (Ming Hui 63, _indica_ rice) by CRISPR/Cas9 system. 18 modern elite _japonica_ rice varieties used for current
rice production in Jiangsu Province were provided by Prof. Jie Yang (Jiangsu Academy of Agricultural Sciences, Nanjing, China). PLANT GROWTH CONDITIONS For paddy field experiments, rice
seedlings were cultivated in Baima field in Nanjing (119° 18’E, 31° 61’N) under long-day conditions (13.5 h day) from May to October. Rice seeds were sown in May and nurtured for 30 days in
seed beds before being transplanted to paddy fields, where they grew until the harvest stage in October. The plant spacing between each seedling was 15 cm, and 7 plants per row × 8 rows for
each replicate of the treatment were planted with at least 3 replicates under LN (150 kg N/ha) or HN (350 kg N/ha) conditions. Urea as the only N source was applied at seedling, tillering
and heading stage, which respectively accounted for 50%, 25% and 25% of the total N. After 3 months of growth, tiller number and yield characters were analyzed in harvest stage. All field
trials were repeated for at least two years. For hydroponic experiments, plants were cultivated in a growth chamber with photocycles of 14 h (30 °C) and 10 h (28 °C), humidity of about 70%,
and photon density of about 200 μmol m−2 s−1. IRRI nutrient solution was used for hydroponics culture17. After 4 days of germination, rice seedling was transferred into 1/4 IRRI nutrient
solution for 3 days, cultured in 1/2 IRRI nutrient solution for 3 days and then in IRRI nutrient solution for 7 days. 0.125 mM and 1.25 mM NH4NO3 were used for LN and HN treatments,
respectively. The nutrient solution was changed daily. At the end of day or at the start of the day, seedling was harvest for phenotype observation or for physiological analyses. GENOME-WIDE
ASSOCIATION ANALYSIS A satisfactory normal distribution of tillering was observed in 120 accessions of MAGIC population in present study. A mixed linear model was employed to conduct
association analysis of the data39. A _q_-value cutoff of 0.05 was used as the threshold to determine the significant association. Horizontal solid line on the Manhattan plot indicates the
genome-wide significance threshold _q_ ≤ 0.05. The Manhattan plot was obtained from the analysis with software TASSEL v5.0 and drawn using the R package40. PLASTID CONSTRUCTION AND PLANT
TRANSFORMATION FOR GENE EDITING All small guide RNA (sgRNA) used for CRISPR/Cas9-facilitated genome editing in this study were designed by CRISPR-PLANT (http://omap.org/crispr/jbrowse/), and
all sgRNA sequences are listed in Supplementary Data 5. All plasmids were transformed into rice via A. tumefaciens strain _EHA105_. The genotypes of transgenic rice lines were confirmed by
DNA sequencing. QUANTITATIVE REVERSE TRANSCRIPTION PCR For the _OsSTP28_ expression assay, blades, sheaths, shoot bases and roots were sampled at the end of night. To determine the rhythmic
expression of _OsSTP28_, the shoot bases of rice seedlings were sampled every 4 h for 2 d. To investigate the response of _OsSTP28_ to N, rice seedlings were cultivated in a hydroponic
system under LN or HN supply for 7 d, and the shoot bases were sampled. For the _OSH15 and GA2ox3/5/8/9_ expression assay, shoot bases were sampled at the end of night. Every sample was
consisted of a mixture of 7 plants. Samples were frozen in liquid nitrogen immediately and well ground, and then total RNA was extracted using Trizol reagent (Cat. #15596018, Thermo Fisher
Scientific, MA, USA) according to the manufacturer’s instructions. RNA concentrations were measured using Nanodrop 2000plus (Thermo Fisher Scientific Inc., MA, USA). cDNA synthesis was
performed using ReverTra Ace®qPCR RT Master Mix with gDNA Remover (Cat. #FSQ-301, TOYOBO LIFE SCI., Shanghai, China). The qRT-PCR reactions were prepared using SYBR Green Master Mix (Cat.
#Q111, Vazyme Bio, Nanjing, China) following the manufacturer’s protocol, and carried out by using a thermal cycler equipped with an QuantStudio™ Real-Time PCR Software system (ABI
QuantStudio3, Thermo Fisher Scientific Inc., MA, USA). Transcript levels were normalized to the _OsActin1_ gene (_Os03g0718100_), and expression values were calculated using the 2-ΔCt method
with the geometric mean of the Ct values of _Actin1_ as a reference. Gene-specific primers are listed in Supplementary Data 5. RNA-SEQ ANALYSIS Shoot bases (∼0.5 cm) of rice from WT and
_stp28-3_ were harvested under LN or HN supply. According to rhythmic fluctuation of glucose, the maximal difference of glucose between wild type and _stp28_ mutant was detected at the end
of night, thus the samples for RNA-seq were harvested at this time point. RNA extraction was performed using the RNeasy Micro Kit (Cat. #74004, Qiagen, Germany). RNA integrity and purity
were assessed using 1% agarose gels to monitor degradation and contamination. RNA purity was checked using the NanoDrop® 2000plus (Thermo Fisher Scientific Inc., MA, USA). RNA integrity was
assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The cDNA libraries were sequenced on the Illumina sequencing platform by BGI
(Shenzhen, China). Clean reads were compared to oryza_sativa_Ensembl_52 (ftp://ftp.ensemblgenomes.org/pub/plants/release-52/fasta/oryza_sativa/dna/). DESeq2 v1.22.1 was used to analyze the
differential expression between the two groups, and the _P_ value was corrected using the Benjamini & Hochberg method. The corrected _P_ value and |log2foldchange| are used as the
threshold for significant difference expression. Data analysis and graphing were conducted by R (ggplot). RNA IN SITU HYBRIDIZATION OF _OSSTP28_ AND HISTOLOGICAL STAINING RNA in situ
hybridizations were carried out as the description by Luo41. 14-old-seedling rice shoot bases were harvested from the hydroponic system at the end of the night and fixed in FAA solution
((Solarbio LIFE SCIENCES, G2355). Thin sections of paraffin-embedded shoot bases of 8–12 μm thickness were generated for the hybridization. A 121 bp-length cDNAs of _OsSTP28_ was selected
for synthesis of digoxigenin (DIG)-labeled RNA sense and antisense probes in vitro by DIG RNA Labeling Kit (SP6/T7) (Cat. #1175025, Roche Applied Science, Basel, Switzerland). The
hybridization reaction was conducted for 24 h at 50 °C. Anti-DIG alkaline phosphatase-conjugated antibody and the NBT (nitro-blue tetrazolium)/ BCIP (5-bromo-4-chloro-3-indolyl phosphate)
staining method were used to visualize the tissue-specific localization of _OsSTP28_ expression in shoot base sections. Photographs were taken by Axio Scope A1 microscope. For histochemical
analysis of _proOsSTP28:GUS_ reporter line, the native promoter sequence (2600 bp) of _OsSTP28_ was amplified from Nipponbare genomic DNA by PCR, linked with the GUS gene to generate
_OsSTP28_ promoter-GUS construct. The construct was then transformed into rice (cv. Nipponbare) via Agrobacterium-mediated transformation. The roots, basal nodes, leaf sheaths and leaf
blades of five independent transgenic rice seedlings were incubated with a GUS reaction mix at 37 °C for 1 h. The samples were decolorized bradytely with anhydrous ethanol, and imaged with
an Olympus BX-51microscope equipped with a DC Vie digital camera. SUBCELLULAR LOCALIZATION ANALYSIS OF OSSTP28 Subcellular localization of OsSTP28 was detected using the full-length cDNA
sequence of _OsSTP28_ fused in-frame into the pSAT6A-EGFP-N1 vector driven by the 35S promoter. Rice protoplasts were isolated from NIP sheaths. Total 10 μg plasmids (pSAT6A-EGFP,
OsRac1-mCherry (a plasma membrane control) and/or STP28-GFP and OsRac1-mCherry) were respectively transformed into rice protoplasts by the polyethylene-glycol-mediated method. Then after
incubated in the dark for 12–15 h at 28 °C, a confocal laser scanning microscope (TCS SP8X, Leica; Germany) was used to observe the fluorescence signals in transformed protoplasts at 488
nm/498–540 nm for eGFP and 552 nm/600–640 nm for mCherry. HETEROLOGOUS EXPRESSION OF _OSSTP28_ IN YEAST AND _X. LAEVIS_ OOCYTES For yeast assay, _OsSTP28_ coding sequence was inserted into
the PDR196 vector, containing the ampicillin-resistance gene, and then transferred into the sugar transporter-deficient yeast strain EBY.VW4000. A single clone of EBY.VW4000 containing
OsSTP28 was precultured on maltose–amino acid medium [0.67% (w/v) yeast nitrogen base, 1% (w/v) casamino acids, 0.002% (w/v) tryptophan and 2% (w/v) maltose] to an A600 of 0.8–1.0, then the
growth of EBY.vw4000 expressing OsSTP28 was assessed on culture media containing 2% concentrations of different sugars (maltose, glucose, fructose, galactose and xylose). Yeast growth
phenotype was recorded after 3 days of cultivation. For oocyte assay, _OsSTP28_ coding sequence (CDS) was cloned into pT7Ts vector. The plasmid was then linearized and purified (Cycle-Pure
Kit, Cat. #D6492, OMEGA). Capped cRNAs of _OsSTP28_ were synthesized using mMESSAGE mMACHINE T7 (Cat. #AM1340; USA), and subsequently, 25–50 ng of cRNAs were injected into healthy-looking
oocytes isolated from Xenopus laevis. The incubation of injected oocytes followed the described method. For glucose efflux assay, 10-20 incubated oocytes were transferred into MBS solution
supplied respectively with 0-, 2-,20-, 40- and 60-mM [13C]-glucose (Shanghai Engineering Research Center of Stable Isotope, CAS: 492-62-6) for 2 h. For glucose influx assay, 0-, 2- and 20-mM
D-glucose was respectively injected into oocytes, and those oocytes was incubated on MBS solution for 2 h. And then, cold MBS buffer was used to wash the oocytes 5 times after incubation at
20 °C, cells were transferred to ice-cold Na-Ringer, washed three times, solubilized with 100 μl 1% (w/v) SDS, and finally measured individually. GLUCOSE, FRUCTOSE AND SUCROSE MEASUREMENTS
To investigate the rhythmic fluctuation of sugar in rice shoot bases, 35-d-old seedlings of WT and _stp28_ mutants were harvested every 4 h for 2 d. To determine sugar response to N, shoot
bases and leaves were respectively stripped from rice seedlings of WT and _stp28_ mutants at the end of the night and/or at the end of the day. Every sample was mixed with 9 plants. All
samples were freeze-dried for 4 days and ground into powder, and then 0.2 g of dry sample was weighed into 10 ml tubes. Subsequently, 2 ml of 80% ethanol was added to each tube, and the
mixture was sonicated at 80 °C for 10 min. The supernatant was absorbed after centrifuge 11,500 g × 5 min. 1 ml of 50% ethanol was added to the precipitate, and the supernatant was taken by
ultrasonic at 80 °C for 10 min and 11,150 g × 5 min. The supernatant was combined into a new centrifuge tube and extracted by adding an equal volume of chloroform. The supernatant was taken
into the new centrifuge tube at 11,150 g × 5 min. Under the condition of 35 °C, the nitrogen blower quickly dried; 50% acetonitrile was added to 1 ml, and then filtered through 0.45 μm
microporous membrane for HPLC determination. The standard curve was prepared with glucose, sucrose and fructose standards. The samples were determination using high performance liquid
chromatography (HPLC) (Waters Associates, MA, USA). ASSAY OF 13C-RADIOLABELING GLUCOSE The fully unfolded leaves of mutants and WT were fed with water containing 2% [13C]-glucose (produced
by Shanghai Engineering Research Center of Stable Isotope, CAS: 492-62-6) for 10 h. The rice was grown in the IRRI nutrient solution (Tang et al., 2012). Subsequently, the leaves, sheaths,
shoot bases, and roots were collected and dried at 105 °C for 1 h. Dry materials were ground into powder and weighed using a Beckman LS650. At least three biological replicates were analyzed
using Isotope Ratio Mass Spectrometry (IRMS, Thermos Fisher Scientific, MA, USA). APOPLASTIC AND INTRACELLULAR SUGAR MEASUREMENTS Plant apoplast space can be divided into air- and
solution-volume. After being treated in different N conditions for 7 days and washed in pure water, samples were weighed as an initial volume (M0). The samples were vacuumed in pure water by
the injector and weighed as M1. Ectoplasmic solution after centrifuge 4500 g × 10 min at 4 °C was collected and weighed as M2. The apoplast space (M0, M1 and M2) was calculated by using a
method of Gentzel42. The concentration of extracted sucrose, glucose and fructose was determined by ultra performance liquid chromatography (UPLC) (Hclass, Waters, USA). To determine the
levels of intracellular sugar, the protoplast solution was isolated by centrifugation. Briefly, after low-speed centrifugation, apoplastic solution was thrown out from the rice tissue. The
remaining rice tissue was cleaned by H2O and dried by airing. After centrifugation at 12,000 g for 20 min, the supernatant was collected for intracellular sugar determination by UPLC
(Hclass, Waters, USA). TRANSIENT TRANSACTIVATION ASSAY IN RICE LEAF PROTOPLAST The full-length _OSH15_ coding sequence was cloned into pGreenII-62-sk vector. ∼3000 bp upstream sequence
(including promoter sequencing and UTR) of _OsGA2ox3/5/8/9_ coding region were amplified as the respective promoter. Rice leaf protoplast of two-week-old seedlings was extracted as a method
described by Zhang17. Each upstream sequence was fused with firefly luciferase (LUC) sequences and introduced to pGreenII-0800 vector to generate the reporter construct. Plasmids containing
the effector and reporter vectors were transfected into rice protoplast. After 12 h of infiltration under dark conditions at 30 °C, protoplast proteins were extracted for the detection of
Renilla luciferase (REN) and LUC activities using the Double-Luciferase Reporter Assay Kit (Cat. #FR201, TransGen Biotech, Beijing, China). Protoplasts without transfection (water only) and
protoplasts transfected with the reporter vector only (empty) were used as negative controls. ELECTROPHORETIC MOBILITY SHIFT ASSAYS Full-length CDS of _OSH15_ was cloned into pET-29a (+)
vector which was transformed into BL21 strain of Escherichia coli to express fusion protein. The promoter sequence of OsGA2ox3/5/8/9 was selected containing about 100 bp up- and down-stream
of the “gatgacctga” motif. Bio-probes were synthesized with biotin labels at both their 3’ and 5’ ends, while the fragment without biotin served as a cold probe. A nondenatured
polyacrylamide gel was prepared and subjected to electrophoresis, and color development was achieved using film. All the primers used for probes and competitors are listed in Supplementary
Data 5. RICE ENDOGENOUS GAS ANALYSIS The shoot bases of NIP and _stp28_ lines were collected at the tillering stage under both LN and HN conditions, immediately frozen in liquid nitrogen,
and ground into powder. The method of determination of endogenous GAs was carried out according to previous reports43. Briefly, plant samples (100 mg FW) and spiked isotope-labeled internal
standards (IS) (D2-GAs, 1 ng) were extracted with 90% MeOH (1 ml) overnight at 4 °C. The extract was then centrifuged at 12,000 g for 15 min at 4 °C and the supernatant was collected. The
crude extracts were loaded onto the connected MCX–WAX cartridges and analyzed by UPLC–MS/MS (Hclass, Waters, USA). DETERMINATION OF GA2-OXIDASES ACTIVITIES GA2-oxidases activities of the
tissues (the same as those used for endogenous GAs analysis) were determined with the GA2-oxidases Kit (HuaBang BIO, HB-P5974Z) following the manufacturer’s instructions. H3K27ME3
MODIFICATION ANALYSIS The ChIP-PCR experiments were carried out as previously described with slight modifications44. Briefly, 2 g of rice shoot bases were collected and ground into a fine
powder using liquid nitrogen. The powdered tissue was incubated with a nuclei isolation buffer for 20 min. Chromatin was fixed using 1% formaldehyde for 10 min, and then the crosslinking
reaction was terminated by adding fresh-prepared glycine. The chromatin was then sonicated using the M220 Focused Ultrasonicator (Covaris, USA) to obtain DNA fragment of ∼350 bp in size.
Immunoprecipitation was performed with 3 μl (1 mg/ml) of anti-H3K27me3 (A2363, ABclonal) at 4 °C overnight. Following this, 30 µl of Magnetic Protein A/G Dynabeads (Invitrogen™, 10002D) were
added, and the mixture was incubated at 4 °C for 4 h. The slurry was subjected to a serial of washes using low salt buffer (150 mM NaCl), high salt buffer (500 mM NaCl), and LiCl wash
buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate). Crosslinks were reversed by incubating the sample at 65 °C overnight. Finally, the
precipitated DNA was purified and quantified by qPCR with the primers listed in Supplementary Data 5. STATISTICAL ANALYSIS Data were collected by Microsoft Excel 2021. All collected data
were analyzed using GraphPad Prism 8 (version 8.3.0). Two-tailed Student’s _t_-test, Dunnett’s multiple test, or Tukey’s HSD test was performed to test the statistical significance, and the
_P_ values of each statistical analysis are noted in figure legends. Graphs were plotted by GraphPad Prism 8 (version 8.3.0) and edited by Adobe Illustrator 2020 (version 24.2.1). ACCESSION
NUMBERS Sequence data from this study can be found in the GeneBank data libraries or The Rice Annotation Project (RAP) under the following accession numbers: _OsSTP28_ (Os11g0594000);
_OsGA2ox3_ (Os01g0757200); _OsGA2ox5_ (Os07g0103500); OsGA2ox8 (Os05g0560900); OsGA2ox9 (Os02g0630300); _OSH15_ (Os07g0129700); _NGR5_ (Os05g0389000); O_sTCP19_ (Os06g0226700); _MOC1_
(Os06g0610350); _OsMADS57_ (Os02g0731200); _OsTB1_ (Os03g0706500); _OsSPL14_ (Os08g0509600); _D14_ (Os03g0203200); _D3_ (Os06g0154200); _D53_ (Os11g0104300); _D27_ (Os11g0587000); _D17_
(Os04g0550600); and _D10_ (Os01g0746400). The accession number of differentially expressed genes shown in Supplementary Fig. 11f, Supplementary Fig. 12b and Supplementary Fig. 18b are listed
in Supplementary Data 6. REPORTING SUMMARY Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY The
RNA-seq data generated in this study have been deposited in the NCBI (National Center for Biotechnology Information) database under accession code PRJNA1070824. The analysis data of RNA-seq
and primers generated in this study are provided in the Supplementary Data 1–5. For any datasets unavailable through the links above, they can be requested from the corresponding author.
Source data are provided with this paper. REFERENCES * Xu, G., Fan, X. & Miller, A. J. Plant nitrogen assimilation and use efficiency. _Annu Rev. Plant Biol._ 63, 153–182 (2012). Article
CAS PubMed Google Scholar * Liu, Q. et al. Improving crop nitrogen use efficiency toward sustainable green revolution. _Annu. Rev. Plant Biol._ 73, 523–551 (2022). Article CAS PubMed
Google Scholar * Hu, B., Wang, W., Chen, J., Liu, Y. & Chu, C. Genetic improvement toward nitrogen-use efficiency in rice: Lessons and perspectives. _Mol. Plant_ 16, 64–74 (2023).
Article CAS PubMed Google Scholar * Guo, J. H. et al. Significant acidification in major Chinese croplands. _Science_ 327, 1008–1010 (2010). Article ADS CAS PubMed Google Scholar *
Sutton, M. A. et al. Too much of a good thing. _Nature_ 472, 159–161 (2011). Article ADS CAS PubMed Google Scholar * Luo, L., Zhang, Y. & Xu, G. How does nitrogen shape plant
architecture? _J. Exp. Bot._ 71, 4415–4427 (2020). Article CAS PubMed PubMed Central Google Scholar * Liu, Y. Q. et al. Genomic basis of geographical adaptation to soil nitrogen in
rice. _Nature_ 590, 600–605 (2021). Article ADS CAS PubMed Google Scholar * Wang, Y. & Li, J. Branching in rice. _Curr. Opin. Plant Biol._ 14, 94–99 (2011). Article CAS PubMed
Google Scholar * Takai, T. Potential of rice tillering for sustainable food production. _J. Exp. Bot._ 75, 708–720 (2023). Article PubMed Central Google Scholar * Luo, L., Pan, S., Liu,
X., Wang, H. & Xu, G. Nitrogen deficiency inhibits cell division-determined elongation, but not initiation, of rice tiller buds. _Isr. J. Plant Sci._ 64, 32–40 (2017). Google Scholar *
Wu, K. et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. _Science_ 367, 6478 (2020). Article Google Scholar * Hou, M. et al. OsPIN9,
an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium. _N. Phytol._ 229, 935–949 (2021). Article CAS Google Scholar * Zhuang, L. et al.
Gibberellic acid inhibition of tillering in tall fescue involving crosstalks with cytokinins and transcriptional regulation of genes controlling axillary bud outgrowth. _Plant Sci._ 287,
110168 (2019). Article CAS PubMed Google Scholar * Liao, Z. et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. _Nat. Commun._ 10, 2738 (2019).
Article ADS PubMed PubMed Central Google Scholar * Meng, L., Guo, L., Ponce, K., Zhao, X. & Ye, G. Characterization of three rice multiparent advanced generation intercross (MAGIC)
populations for quantitative trait loci identification. _Plant Genome_ 9, 2 (2016). Article Google Scholar * Wang, F. et al. The rice circadian clock regulates tiller growth and panicle
development through strigolactone signaling and sugar sensing. _Plant Cell_ 32, 3124–3138 (2020). Article CAS PubMed PubMed Central Google Scholar * Zhang, S. et al. Nitrogen mediates
flowering time and nitrogen use efficiency via floral regulators in rice. _Curr. Biol._ 31, 671–683 (2021). Article CAS PubMed Google Scholar * Marchive, C. et al. Nuclear retention of
the transcription factor NLP7 orchestrates the early response to nitrate in plants. _Nat. Commun._ 4, 1713 (2013). Article ADS PubMed Google Scholar * Hu, B. et al. Nitrate-NRT1.1B-SPX4
cascade integrates nitrogen and phosphorus signalling networks in plants. _Nat. Plants_ 5, 401–413 (2019). Article CAS PubMed Google Scholar * Zhao, H. et al. An inferred functional
impact map of genetic variants in rice. _Mol. Plant._ 14, 1584–1599 (2021). Article CAS PubMed Google Scholar * Deng, X. et al. A novel insight into functional divergence of the MST gene
family in rice based on comprehensive expression patterns. _Genes_ 10, 239 (2019). Article CAS PubMed PubMed Central Google Scholar * Schleucher, J., Vanderveer, P. J. & Sharkey,
T. D. Export of carbon from chloroplasts at night. _Plant Physiol._ 118, 1439–1445 (1998). Article CAS PubMed PubMed Central Google Scholar * Li, X. et al. Control of tillering in rice.
_Nature_ 422, 618–621 (2003). Article ADS CAS PubMed Google Scholar * Guo, S. Y. et al. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. _Nat. Commun._
4, 1566 (2013). Article ADS PubMed Google Scholar * Takeda, T. et al. The OsTB1 gene negatively regulates lateral branching in rice. _Plant J._ 33, 513–520 (2003). Article CAS PubMed
Google Scholar * Jiao, Y. et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. _Nat. Genet._ 42, 541–544 (2010). Article CAS PubMed Google Scholar * Lo, S.
F. et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. _Plant Cell_ 20, 2603–2618 (2008). Article CAS PubMed PubMed Central
Google Scholar * Ye, R. et al. Glucose-driven TOR-FIE-PRC2 signalling controls plant development. _Nature_ 609, 986–993 (2022). Article ADS CAS PubMed PubMed Central Google Scholar *
Xu, L. & Shen, W. H. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. _Curr. Biol._ 18, 1966–1971 (2008). Article CAS PubMed Google Scholar * Zhao, L.
et al. Integrative analysis of reference epigenomes in 20 rice varieties. _Nat. Commun._ 11, 2658 (2020). Article ADS CAS PubMed PubMed Central Google Scholar * Fu, L. Y. et al.
ChIP-Hub provides an integrative platform for exploring plant regulome. _Nat. Commun._ 13, 3413 (2022). Article ADS CAS PubMed PubMed Central Google Scholar * Sasaki, A. et al. Green
revolution: a mutant gibberellin-synthesis gene in rice. _Nature_ 416, 701–702 (2002). Article ADS CAS PubMed Google Scholar * Spielmeyer, W. et al. Isolation of gibberellin metabolic
pathway genes from barley and comparative mapping in barley, wheat and rice. _Theor. Appl. Genet._ 109, 847–855 (2004). Article CAS PubMed Google Scholar * Bolduc, N. & Hake, S. The
maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene _ga2ox1_. _Plant Cell_ 21, 1647–1658 (2009). Article CAS PubMed PubMed Central Google Scholar *
Wang, B., Smith, S. M. & Li, J. Genetic regulation of shoot architecture. _Annu. Rev. Plant Biol._ 69, 437–468 (2018). Article CAS PubMed Google Scholar * Li, M. et al. Knockout of
the sugar transporter OsSTP15 enhances grain yield by improving tiller number due to increased sugar content in the shoot base of rice (Oryza sativa L.). _N. Phytol._ 241, 1250–1265 (2024).
Article CAS Google Scholar * Lemoine, R. et al. Source-to-sink transport of sugar and regulation by environmental factors. _Front. Plant Sci._ 4, 272 (2013). Article CAS PubMed PubMed
Central Google Scholar * Slewinski, T. L. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. _Mol. Plant_ 4,
641–662 (2011). Article CAS PubMed Google Scholar * Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. _Nat. Genet._ 44, 821–824 (2012).
Article CAS PubMed PubMed Central Google Scholar * Bradbury, P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. _Bioinformatics_ 23, 2633–2635
(2007). Article CAS PubMed Google Scholar * Luo, L. et al. Developmental analysis of the early steps in strigolactone-mediated axillary bud dormancy in rice. _Plant J._ 97, 1006–1021
(2019). Article CAS PubMed PubMed Central Google Scholar * Gentzel, I., Giese, L., Zhao, W., Alonso, A. & Mackey, D. A simple method for measuring apoplast hydration and collecting
apoplast contents. _Plant Physiol._ 179, 1265–1272 (2019). Article CAS PubMed PubMed Central Google Scholar * Xin, P., Guo, Q., Li, B., Cheng, S., Yan, J. & Chu, J. A tailored
high-efficiency sample pretreatment method for simultaneous quantification of 10 classes of known endogenous phytohormones. _Plant Commun._ 1, 100047 (2020). Article PubMed PubMed Central
Google Scholar * Xu, Y. et al. Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. _Nucleic Acids Res._ 42, 10960–10974 (2014).
Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank Prof. Yufeng Wu and Dr. Le Luo (Nanjing Agricultural University) for the help with GWAS
analysis and RNA in situ hybridization, Prof. Jie Yang (Jiangsu Academy of Agricultural Science) for providing modern elite _japonica_ rice varieties, Prof. Bin Hu (South China Agricultural
University) for providing _tcp19-1_ and _tcp19-2_ rice mutants, Prof. Mingjun Li (Northwest A&F University) for providing yeast strain EBY.VW4000 and Dr. Fengying Duan (Chinese Academy
of Agricultural Sciences) for the technical help with the measurement of apoplastic sugars, Dr. Dijun Chen and Tao Zhu (Nanjing University) for the help of remining the epigenome data in
rice. This study is supported by National Key Research and Development Program of China (2021YFF1000400), National Natural Science Foundation of China (31930101), Jiangsu Provincial Key
Research and Development Program (BE2020339), Jiangsu Seed Industry Revitalization Project (JBGS [2021] 011) with grants to G.X., and by Jiangsu Provincial Key Research and Development
Program (BE2022336) with a grant to Y.L. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Nanjing Agricultural
University, Nanjing, 210095, China Jinfei Zhang, Yuyi Zhang, Mengfan Xu, Xinyu Guan, Shunan Zhang, Hongye Qu, Mian Gu, Ying Liu & Guohua Xu * Key Laboratory of Plant Nutrition and
Fertilization in Low-Middle Reaches of the Yangtze River, Ministry of Agriculture, Nanjing Agricultural University, Nanjing, 210095, China Jinfei Zhang, Yuyi Zhang, Mengfan Xu, Xinyu Guan,
Shunan Zhang, Hongye Qu, Mian Gu, Ying Liu & Guohua Xu * School of Agriculture, Shenzhen Campus of Sun Yat-sen University, Shenzhen, 518107, China Jingguang Chen * College of Life
Sciences, Nanjing Agriculture University, Nanjing, 210095, China Cui Wu & Yifeng Xu * National Center for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology,
Chinese Academy of Sciences, Beijing, 100101, China Jinfang Chu * University of Chinese Academy of Sciences, Beijing, 100049, China Jinfang Chu Authors * Jinfei Zhang View author
publications You can also search for this author inPubMed Google Scholar * Yuyi Zhang View author publications You can also search for this author inPubMed Google Scholar * Jingguang Chen
View author publications You can also search for this author inPubMed Google Scholar * Mengfan Xu View author publications You can also search for this author inPubMed Google Scholar * Xinyu
Guan View author publications You can also search for this author inPubMed Google Scholar * Cui Wu View author publications You can also search for this author inPubMed Google Scholar *
Shunan Zhang View author publications You can also search for this author inPubMed Google Scholar * Hongye Qu View author publications You can also search for this author inPubMed Google
Scholar * Jinfang Chu View author publications You can also search for this author inPubMed Google Scholar * Yifeng Xu View author publications You can also search for this author inPubMed
Google Scholar * Mian Gu View author publications You can also search for this author inPubMed Google Scholar * Ying Liu View author publications You can also search for this author inPubMed
Google Scholar * Guohua Xu View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS G.X., Y.L., and J.Z. conceived the project and designed the
experiments. J.Z. performed most of the experiments and data analysis, Y.Z. helped with the biochemistry experiments. J.C., M.X., and X.G. helped with the field experiments, C.W. and Y.X.
performed ChIP-PCR assay to detect H3K27me3 modification, J.C. helped with endogenous GAs measurement, S.Z., H.Q., M.G. helped with data analysis and graphs plotting. G.X., Y.L., and J.Z.
wrote the manuscript with the support from all authors. All authors contributed to the finalization of the manuscript. CORRESPONDING AUTHORS Correspondence to Ying Liu or Guohua Xu. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Communications_ thanks Jian Jin, Zuofeng Zhu and the other,
anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with
regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTION OF ADDITIONAL SUPPLEMENTARY
FILES SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA 3 SUPPLEMENTARY DATA 4 SUPPLEMENTARY DATA 5 SUPPLEMENTARY DATA 6 REPORTING SUMMARY SOURCE DATA SOURCE DATA FIGURE1 SOURCE
DATA FIGURE2 SOURCE DATA FIGURE3 SOURCE DATA FIGURE4 SOURCE DATA FIGURE5 SOURCE DATA FIGURE6 SOURCE DATA SUPPLEMENTARY FIG.1 SOURCE DATA SUPPLEMENTARY FIG.2 SOURCE DATA SUPPLEMENTARY FIG.4
SOURCE DATA SUPPLEMENTARY FIG.5 SOURCE DATA SUPPLEMENTARY FIG.7 SOURCE DATA SUPPLEMENTARY FIG.8 SOURCE DATA SUPPLEMENTARY FIG.9 SOURCE DATA SUPPLEMENTARY FIG.10 SOURCE DATA SUPPLEMENTARY
FIG.11 SOURCE DATA SUPPLEMENTARY FIG.12 SOURCE DATA SUPPLEMENTARY FIG.13 SOURCE DATA SUPPLEMENTARY FIG.14 SOURCE DATA SUPPLEMENTARY FIG.16 SOURCE DATA SUPPLEMENTARY FIG.17 SOURCE DATA
SUPPLEMENTARY FIG.18 SOURCE DATA SUPPLEMENTARY FIG.19 SOURCE DATA SUPPLEMENTARY FIG.20 SOURCE DATA SUPPLEMENTARY FIG.21 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a
Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as
long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not
have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s
Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Zhang, J., Zhang, Y., Chen, J. _et al._ Sugar transporter modulates
nitrogen-determined tillering and yield formation in rice. _Nat Commun_ 15, 9233 (2024). https://doi.org/10.1038/s41467-024-53651-1 Download citation * Received: 15 February 2024 * Accepted:
16 October 2024 * Published: 25 October 2024 * DOI: https://doi.org/10.1038/s41467-024-53651-1 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content:
Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
