Hypomethylation of proximal cpg motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT AIM: The promoter of human interleukin-10 (IL10), a cytokine crucial for suppressing inflammation and regulating immune responses, contains an interspecies-conserved sequence with
CpG motifs. The aim of this study was to investigate whether methylation of CpG motifs could regulate the expression of _IL10_ in rheumatoid arthritis (RA). METHODS: Bioinformatic analysis
was conducted to identify the interspecies-conserved sequence in human, macaque and mouse _IL10_ genes. Peripheral blood mononuclear cells (PBMCs) from 20 RA patients and 20 health controls
were collected. The PBMCs from 6 patients were cultured in the presence or absence of 5-azacytidine (5 μmol/L). The mRNA and protein levels of _IL10_ were examined using RT-PCR and ELISA,
respectively. The methylation of CpGs in the _IL10_ promoter was determined by pyrosequencing. Chromatin immunoprecipitation (ChIP) assays were performed to detect the cyclic AMP response
element-binding protein (CREB)-DNA interactions. RESULTS: One interspecies-conserved sequence was found within the _IL10_ promoter. The upstream CpGs at −408, −387, −385, and −355 bp were
hypermethylated in PBMCs from both the RA patients and healthy controls. In contrast, the proximal CpG at −145 was hypomethylated to much more extent in the RA patients than in the healthy
controls (_P_=0.016), which was correlated with higher _IL10_ mRNA and serum levels. In the 5-azacytidine-treated PBMCs, the CpG motifs were demethylated, and the expression levels of _IL10_
mRNA and protein was significantly increased. CHIP assays revealed increased phospho-CREB binding to the _IL10_ promoter. CONCLUSION: The methylation of the proximal CpGs in the _IL10_
promoter may regulate gene transcription in RA. SIMILAR CONTENT BEING VIEWED BY OTHERS DNA METHYLATION OF IFI44L AS A POTENTIAL BLOOD BIOMARKER FOR CHILDHOOD-ONSET SYSTEMIC LUPUS
ERYTHEMATOSUS Article Open access 21 March 2024 CORRELATION ANALYSIS OF CIRCULATING _PCDH17_ DNA METHYLATION CHANGES WITH RHEUMATOID ARTHRITIS PATIENTS Article Open access 23 May 2025
GENE-REGULATORY NETWORK ANALYSIS OF ANKYLOSING SPONDYLITIS WITH A SINGLE-CELL CHROMATIN ACCESSIBLE ASSAY Article Open access 10 November 2020 INTRODUCTION Rheumatoid arthritis (RA) is an
inflammatory and autoimmune disease1. Proinflammatory and anti-inflammatory cytokines are involved in the initiation and progression of RA; therefore, a detailed study of the regulatory
mechanism of the production of cytokines during the development of RA is of utmost importance. Interleukin-10 (IL10), produced mainly by monocytes, T helper type 2 (Th2) and regulatory T
cells (Treg), plays a crucial role in suppressing inflammation and regulating the immune response2. It contributes to the growth and differentiation of B cells, but inhibits T cell
proliferation. IL10 can fight inflammation by down-regulating the production of proinflammatory cytokines, such as tumor necrosis factor alpha (TNFα), interleukin-1 beta (IL1β),
interleukin-6 (IL6), and others; it also stimulates the production of cytokine inhibitors (_eg_, the IL1 receptor antagonist and the soluble TNF receptor). Evidence has indicated that IL10
is involved in the pathogenesis of RA. Several murine models of RA are markedly exacerbated in IL10-deficient mice3, 4. Exogenous IL10 can prevent the development of arthritis and regulate
immune cell function5, 6. The _IL10_ gene exhibits substantial polymorphism in the promoter region that correlates with transcription. Recent studies have found that the methylation status
of CpG sites is related to cytokine expression7. Therefore, the extent of methylation at CpG sites correlates with the level of cytokine production. Furthermore, the amount of methylation at
CpG sites has been shown to be related to cells' differentiation or activation. Several CNS regions in the _IL10_ gene have been identified by bioinformatic analysis, including the
5′-proximal region, promoter, and introns8, and the hypomethylation of CpGs around intron 4 may enhance the expression of _IL10_9. However, a correlation was not found between the CpG
methylation status of the _IL10_ promoter and IL10 production. _IL10_'s promoter region contains many CpGs and putative transcription factors binding sites, such as Sp-1, CCAAT/enhancer
binding protein (C/EBP-β) and cyclic AMP response element-binding protein (CREB). Several papers reported that the activated cAMP/CREB signaling pathway could enhance the IL10 production in
human monocytes, and IFN-gamma suppressed IL10 synergize by regulating CREB/AP-1 protein expression10, 11. CREB is expressed at a higher level in the synovial cells of patients with RA, and
it is implicated in IL6 production from synovial cells12. However, whether the methylation status of a single CpG site in the _IL10_ promoter can regulate its transcription through
affecting CREB's binding to this region in RA remains unclear. Some epigenetic clues to RA have been revealed. Global hypomethylation of the T cell genome13 and methylation of the death
receptor 3 gene (_DR3_)14 and the _IL6_ promoter are related to the pathogenesis of RA15. Because epigenetic alterations are reversible, they may provide new molecular targets for
therapeutic intervention. Therefore, by bioinformatics analysis, we analyzed the human _IL10_ gene to seek CNS and CRE. Then, we investigated whether methylation of the _IL10_ promoter could
regulate its expression. Furthermore, we studied whether DNA methylation had an effect on CREB binding to the promoter, thus illustrating the mechanism underlying the regulatory effects of
DNA methylation on gene expression. MATERIALS AND METHODS PATIENTS AND CONTROLS Peripheral blood from 20 patients with active RA and 20 healthy controls (HCs) were studied. RA patients
recruited from the Second and Third Affiliated Hospitals of Hebei Medical University (Shijiazhuang, China) were in conformity with the American College of Rheumatology Criteria16. Research
ethics approval was obtained from Hebei Medical University Research Ethics Committee (Shijiazhuang, China), and informed consent was obtained from individual patients. Healthy controls were
collected from Hebei Province Blood Center. Of the RA patients, 4 were male, and 16 were female with their mean±SD age being 39.6±11.4 years old (ranging from 20 to 58 years old). Most of
the RA patients in this study were in moderate disease activity. The mean DAS28 was 4.38±1.8 (range 1.9–8.8), the mean time from onset was 25.33±22.33 months (ranging from 3 months to 6
years), the mean erythrocyte sedimentation rate (ESR) was 37.4±28.13 mm/h (range 6–123), the mean C-reactive protein (CRP) was 17.27±20.63 mg/L (range 1.96–92.9), and the mean rheumatoid
factor (RF) was 164.16±122.92 U/mL (range 17.2–447). Of the healthy individuals, 4 were male, and 16 were female. Their mean±SD age was 38.4±10.2 years old (ranging from 18 to 56 years old).
PERIPHERAL BLOOD MONONUCLEAR CELLS (PBMCS) AND CELL CULTURE PBMCs were isolated by Ficoll-Hypaque centrifugation from peripheral blood of RA patients and HCs. PBMCs from six patients and
controls were cultured in RPMI-1640 medium supplemented with 25 mmol/L HEPES, 4 mmol/L _L_-glutamine, 100 units/mL penicillin/streptomycin, and 5% heat-inactivated fetal calf serum with 1
μg/mL phytohemagglutinin (PHA; Sigma, USA) and 10 μg/mL lipopolysaccharides (LPS; Sigma, USA) . In the presence or absence of 5 μmol/L 5-azacytidine (5-azaC) (Sigma, USA) for 72 h, 2×106
cells/well were cultured according to Mi's method17. MRNA AND PROTEIN EXPRESSION After being extracted from PBMCs using TRIzol reagent (SBS, China), the RNA was reverse transcribed in
the presence of 1×PCR buffer, 1 mmol/L dNTPs, 20 units AMV reverse transcriptase, 20 units RNA guard ribonuclease inhibitor and 2.5 mmol/L random primers (SBS, China) in a final reaction
volume of 20 μL. Reactions were conducted at 30 °C for 10 min and 42 °C for 60 min using a PTC-200 PCR system (MJ Research, USA). The level of _IL1_0 mRNA transcripts was determined by
semiquantitative reverse transcription polymerase chain reaction (RT-PCR). β-actin was used as the internal standard for each RT-PCR. TNFα was measured as a control for cytokine expression.
The specific primers used for the amplification of _IL10_ were the following: forward, 5′-TCAGGGTGGCGACTCTAT-3′, and reverse, 5′-TGGGCTTCTTTCTAAATCGTTC-3′; β-actin: forward,
5′-CATCCTGCGTCTGGACCT-3′, and reverse, 5′-TCAGGAGCAATGATCTTG-3′; TNFα: forward, 5′-CGAGTCTGGGCAGGTCTA-3′, and reverse, 5′-GTGGTGGTCTTGTTGCTTAA-3′. PCR amplification was conducted on a
PTC-200 PCR system using 5 μL cDNA, 1×GeneAmp PCR Gold buffer (Applied Biosystems), 1.5 mmol/L MgCl2 (Applied Biosystems), 200 μmol/L dNTPs (Shanghai Sangon Biological Engineering
Technology, Shanghai, China), 0.6 μmol/L forward and reverse primers, 1 unit AmpliTaq Gold DNA polymerase (Applied Biosystems) in a 20 μL total reaction volume. The PCR amplification
conditions were denaturation for 3 min at 94 °C; amplification for 30 cycles of 45 s at 94 °C, 45 s at 50 °C for _IL10_ (or 58 °C for β-actin, 55 °C for TNFα), and 45 s at 72 °C; and
extension for 10 min at 72 °C. PCR products were resolved by 2% agarose gel electrophoresis with ethidium bromide. The images were recorded and quantified by Hema analyzer (Udine, Italy).
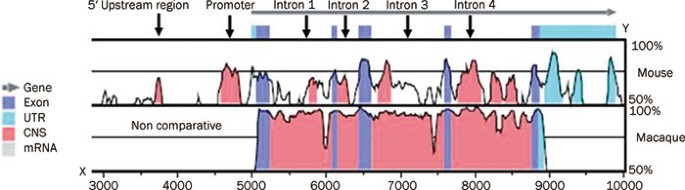
The level of IL10 in serum was measured by ELISA (Bender MedSystems, Austria) according to the manufacturer's instructions. BIOINFORMATICS Alignments between mouse and human _IL10_
loci, and between macaque and human _IL10_ loci were performed, and the extent of DNA sequence homology was computed with a web-based program called Regulatory Visualization Tools for
Alignment (rVISTA; http://www.gsd.lbl.gov/vista)18. The plot of the percentage of sequence identity referred to the human sequence. Regions with a length of at least 100 bp, which showed at
least 75 percent sequence identity at each segment of the alignment between successive gaps, are identified as CNS and are shown in Figure 1. DNA METHYLATION STATUS DNA was extracted from
the PBMCs by the phenol-chloroform method. The DNA concentration was measured by a spectrophotometer (Bio-Rad, USA). One microgram of DNA was treated with sodium bisulphite as previously
described19. PCR and pyrosequencing primers were designed using PSQ Assay Design software (Biotage, Sweden) to amplify the CpG dinucleotides in the _IL10_ promoter. The primer sequences used
are in Table 1. PCR was performed in a 50 μL reaction system of GeneAmp 9700 (Applied Biosystems, USA). Each PCR reaction contained 50 ng bisulfite treated DNA, 1×PCR buffer, 2.5 mmol/L
MgCl2, 0.2 mmol/L dNTPs, 0.2 μmol/L forward primer, 0.2 μmol/L reverse primer, and 1 unit AmpliTaq Gold (Applied Biosystems, USA). The PCR conditions were initiated with a heated lid at 95
°C for 5 min, followed by 50 cycles of 95 °C for 15 s, 51 °C for 30 s, 72 °C for 30 s, and, finally, 72 °C for 5 min. PCR product quality verification was performed using 2% agarose gels
with ethidium bromide. Fifty microliters of PCR products was used for each pyrosequencing reaction. Pyrosequencing methylation analysis was conducted by the Pyro Q-CpG system (PyroMark ID,
Biotage, Sweden) according to the manufacturer's protocol. In brief, the PCR product was bound to streptavidin-coated Sepharose beads (GE Healthcare Bio-sciences AB, Sweden). The
Sepharose beads containing the immobilized PCR product were purified in 70% ethanol for 5 s, denatured in denaturing buffer (Biotage) for 5 s, and washed with washing buffer (Biotage) for 10
s using the pyrosequencing Vacuum Prep Tool (Biotage). Then, 0.5 μmol/L sequence primer was annealed to the purified, single-stranded PCR product, and pyrosequencing was fulfilled by the
Pyro Q-CpG system. The level of methylation for each cytosine locus on CpG sites was expressed as the percentage of mC/(mC+C) (mC is methylated cytosine, C is unmethylated cytosine). Non-CpG
cytosine residues were used as controls to verify bisulfite conversion. CHROMATIN IMMUNOPRECIPITATION (CHIP) ASSAY According to the manufacturer's instructions, the ChIP assay was
performed to detect phospho-CREB binding to the _IL10_ promoter in the PBMCs of patients _in situ_ before and after 5-azaC treatment. ChIP was performed with the Chromatin
Immunoprecipitation Assay Kit (Upstate Biotechnology, No 17-295) and anti-phospho-CREB antibody (Ser133) (Upstate Biotechnology, No 06-519). PCR was performed on _IL10_'s upstream
promoter and proximal promoter, and an intron was amplified as internal control. Primer sequences were upstream promoter primer sequence, forward, 5′-GAAGTCTTGGGTATTCATCC-3′, reverse,
5′-GCTGTGGGTTCTCATTCG-3′; promoter primer sequence, forward, 5′-CTCCCCAGGAAATCAACT-3′, reverse, 5′-AAAAGCCACAATCAAGGT-3′; intron primer sequence, forward, 5′-TTAGAGCGTTTCCAGACC-3′, reverse,
5′-ACCTATGTCAACCCTTCG-3′. PCR was performed in a 20 μL reaction in GeneAmp 9700 (Applied Biosystems, USA). Each PCR reaction contained 100 ng DNA, 1×PCR buffer, 2.0 mmol/L MgCl2, 0.2 mmol/L
dNTPs, 0.2 μmol/L forward and reverse primers, and 1 unit AmpliTaq Gold (Applied Biosystems, USA). The PCR conditions were initiated with a heated lid at 95 °C for 3 min, followed by 35
cycles of 94 °C for 45 s, 51–55 °C for 45 s, 72 °C for 45 s, and, finally, 72 °C for 5 min. PCR products were analyzed in an agarose gel. Values were normalized according to the input.
STATISTICAL ANALYSIS All calculations were performed on a Microsoft computer using SPSS software (version 14.0). Data were analyzed by independent-samples _t_ test and paired _t_ test.
Correlation coefficients were calculated by Pearson rank correlation (_r_) and Spearman rank correlation where applicable. RESULTS BIOINFORMATIC APPROACH TO THE IDENTIFICATION OF CRES IN
IL10 LOCUS We performed a bioinformatic search for CREs in the _IL10_ locus using web-based software, rVISTA. This program can identify interspecies-conserved sequences for specific
transcription factors by linking to the most widely used database, TRANSFAC. As shown in Figure 1, four CREs were found between macaques and humans ranging from introns 1 to 4 and all were
interspecies-conserved. As the readily available macaque IL10 sequence does not contain the upstream promoter region, we did not compare the IL10 promoter region of macaque with that of
humans. Three CREs were found between mice and humans, including the 5′-proximal region, promoter and introns 1 to 4, and the interspecies-conserved CRE was in the promoter and a short
fragment of intron 4. This suggested that the CRE in the promoter, designated as p-CRE, may be important for the regulation of _IL10_ gene expression. METHYLATION STATUS OF _IL10_ PROMOTER
There are at least five CpG sites in the _IL10_ promoter region, four located at −408, −387, −385, −355 bp (upstream) and one at −145 bp (proximal) relative to the transcription start site.
It was found that methylation of individual CpG sites was similar in PBMCs from the 20 RA patients and the 20 HCs; that is, the upstream CpG motifs were hypermethylated, while the proximal
CpG motif was hypomethylated. However, in the patients, methylation of cytosine at −145 bp was significantly less than that in the HCs (18%±7% _versus_ 33%±6.6%; _P_=0.016) (Figure 2).
METHYLATION STATUS AND MRNA EXPRESSION We then studied the relationship between the methylation status of the _IL10_ gene and mRNA expression. The mRNA (0.89±0.21 _versus_ 0.70±0.13,
_P_=0.009) and serum level (71.36±43.25 pg/mL _versus_ 42.85±2.99 pg/mL, _P_=0.012) of IL10 was significantly higher in the patients than in HCs. Furthermore, IL10 mRNA levels of the
patients were correlated with the RA factor (_r_=0.526, _P_=0.016). As for control cytokine, TNFα mRNA expression was significantly higher in the PBMCs of RA patients than in controls
(0.5143±0.5249 _versus_ 0.0861±0.1434, _P_=0.010). Compared with the 20 HCs, the methylation frequency of −145C was significantly lower in the 20 RA patients, which resulted in inversely
higher _IL10_ mRNA levels (Figure 3). Therefore, hypomethylation of −145C was correlated with higher mRNA expression (_r_=-0.746, _P_=0.001). Furthermore, after PBMCs from the six patients
were cultured in the presence of 5-azaC, all five CpG motifs were demethylated (Table 2), and the mRNA of _IL10_ (0.2866±0.15 _versus_ 0.4488±0.13, _P=0.037_) and protein levels (5.30±8.48
pg/mL _versus_ 72.00±7.12 pg/mL, _P_=0.012) were increased in the presence of 5-azaC, whereas the mRNA level of β-actin and TNFα remained constant. IL10 mRNA (1 _versus_ 1.7084±1.8361,
_P_=0.47) and protein expressions (1.4349±0.5887 pg/mL _versus_ 3.275±2.0533 pg/mL, _P_=0.093) were increased after the control cells were treated with 5-azaC, but the difference was not
significant. EFFECTS OF METHYLATION AT IL10 CPG MOTIFS ON THE BINDING OF A TRANSCRIPTION FACTOR There are at least three possible binding sites for CREB and C/EBP family members in the
_IL10_ promoter: one in upstream sequences at −408C and the others upstream of the promoter (−292 to −304 bp) or in the promoter region (−49 to −59 bp). ChIP assay results showed that the
density of the _IL10_ promoter and upstream promoter bands was significantly higher in the 5-azaC treated group than in the untreated group (Figure 4). DISCUSSION In many murine models of
RA, IL10 attenuated arthritis5, 6; however, the clinical improvement of RA patients was unstable20. Gene therapies have been attempted to overcome this defect. The treatment of a single
joint by local delivery of the viral _IL10_ gene may protect multiple joints of the same mice with CIA21. With regard to IL10 production, Im SH _et al_ analyzed the _IL10_ gene and showed
that the promoter was highly conserved between humans and mice22. In the present study, we found one interspecies-conserved CRE within the promoter as well. Dong _et al_8 analyzed the CpG
methylation pattern of the _IL10_ locus and detected two slightly demethylated CpGs in the proximal promoter in IL10+ _vs_ IL10− Th cells. They concluded that there was no correlation
between methylation pattern and _IL10_ expression. In contrast, our study revealed a significant methylation of p-CRE for IL10 production. Although we observed that upstream CpG motifs were
highly methylated in both the patients and the HCs, the methylation levels of the −145 CpG site were lower than that of upstream CpG motifs in the patients and HCs and were much lower in the
patients, whose _IL10_ mRNA and serum levels were elevated. Furthermore, the lower methylation of −145C correlated with higher mRNA levels in RA. Thus, the hypomethylation of −145C may
regulate gene expression at the transcriptional level and be responsible for the development of RA in some people. IL10 is produced mainly by monocytes, B cells, Th2, and Treg. PBMCs are a
mixture of a variety of cells, including T cells, B cells, and monocytes. The cells in the peripheral blood interregulate each other's function through their secreted cytokines, which
form a complicated network. Measuring the mRNA level of IL10 in PBMCs and the protein level in serum could reflect the general IL10 expression status and the inter-regulation among cells. In
this study, we detected higher IL10 mRNA and protein levels in RA patients than in controls, which may be related to patients' varied disease activities and treatment statuses, and
this finding is consistent with other reports in the field23, 24, 25. For example, Iwata _et al_23 reported that the mean B10 + progenitor B10 (B10pro) cell frequencies were significantly
higher in patients with RA compared with healthy controls. Additionally, IL10 production by blood B cells has been reported to be higher in patients with RA24. Sempere-Ortells _et al_25
found increased IL10 and a higher proportion of CD4(+)IL10(+), CD4(+)CD25(int)IL10(+) cells in peripheral blood of patients with moderately active disease. Taken together, these findings
suggested that during active disease, these cell subsets produce progressively larger amounts of IL10 to regulate inflammation in response to the activity of the disease. The demethylation
of −145 CpG in the _IL10_ promoter could elevate its mRNA expression, which may ameliorate the symptoms of RA. In this paper, we completed a preliminary study of the IL10 expression and
promoter methylation status from the peripheral blood of RA patients. Further study is needed on the regulation mechanisms of all IL10-secreting cells after the separation and purification
of individual cell lineages. Nile _et al_15 found that the differences identified between RA patients and healthy individuals could either be a primary phenomenon, implying that lower
methylation at _IL6_ −1099C is inversely correlated with development of RA or are secondary to the disease process or treatment. It was found in our study that lower methylation of _IL10_
−145C might lead to similar results. Medications such as corticosteroids can alter lymphocyte trafficking and cause relative lymphopenia26. It is increasingly recognized that glucocorticoids
change chromatin structure27. Glucocorticoids may lead to the deacetylation of histones, resulting in the tighter coiling of DNA and reduced access of transcription factors to their binding
sites, thereby suppressing the activity of DNA methyltransferases. Methotrexate therapy has been associated with increased DNA methylation28. Participants in our study had not used any of
these treatments or other immunosuppressants for 3 months before enrollment in the study. Ibuprofen, nimesulide and diclofenac sodium enteric-coated tablets were used to help patients reduce
inflammation and pain, and these therapies have no effect on DNA methylation. DNA methyltransferase activity is proliferation dependent and cell cycle regulated29, 30. Thus, any therapy
that affects proliferation and interferes with DNA methyltransferase activity may affect the status of DNA methylation, but further study is needed to confirm this. Prospective studies of
patients with recent-onset RA will be required to elucidate the potential affects of drugs on DNA methylation. Epigenetic alterations are reversible, providing new molecular targets for
therapy. 5-azaC is an inhibitor of DNA methyltransferases. In this study, 5-azaC induced demethylation of the upstream and proximal CpG sites, and _IL10_ mRNA expression increased in PBMCs
of the patient. Thus, this experiment may contribute to overcoming the relative deficiency of IL10 in RA patients and be hopeful for changing RA progression. To test whether methylation of
the CpG motifs can inhibit transcription factor binding, we performed ChIP assays. Several transcription factors, such as GRE, CREB, Sp1, and C/EBPs, can bind to the _IL10_ promoter. It was
reported that cAMP/CREB signaling was involved in _IL10_ transcription in human monocytic cells11, and C/EBP and CREB/ATF were activated as well. CREB, a member of the leucine zipper family
of transcription factors, can be activated by phosphorylation at serine residue 133 (Ser-133) and further induces transcription of genes in response to the cAMP pathway. C/EBPs, another
member of the leucine zipper family, contain at least six members (C/EBP alpha to C/EBP zeta). C/EBPβ has been shown to interact with CREB/ATF31. Therefore, an antibody specific for
phospho-CREB can be combined with phospho-CREB-DNA and the CREB/ATF-C/EBPβ-DNA compound. Our results indicated that 5-azaC treatment increased CREB and/or C/EBPβ binding to the region
upstream of the promoter and to the promoter region, which was consistent with demethylated CpGs and increased mRNA expression. −408C was contained in CREB and C/EBPβ binding sequences, and
the result indicated its methylation inhibited CREB and C/EBPβ binding to the upstream promoter directly. All other CpG motifs were also demethylated (Table 1). Therefore, simultaneous
demethylation of upstream and proximal CpG motifs in the _IL10_ promoter may allow easier access for transcription factors to their binding sites, thus enhancing _IL10_ transcription. Other
transcription factors, aside from CREB, may participate in the regulation of the IL10 gene, such as STAT332, another transcription factor that binds near −145, NF-κB, and GRE. Whether the
other transcription factors' binding activities are regulated by DNA methylation remains to be investigated. Until now, there have been very limited data regarding the epigenetic
regulation of genes in RA; only the methylation status of _DR3_14 and _IL6_15 was reported. The histone deacetylase activity in synovial tissues from patients with RA was approximately
2-fold lower than that in synovial tissues from patients with osteoarthritis or from normal controls33. A single intravenous injection of depsipeptide (FK228), a histone deacetylase
inhibitor, inhibited joint swelling, synovial inflammation and subsequent bone and cartilage destruction in mice with autoantibody-mediated arthritis (AMA)34. The authors also found that
intravenous treatment with FK228 induced the hyperacetylation of histone H3 and H4 in synovial cells. Furthermore, histone deacetylase inhibitor, trichostatin A (TSA), in combination with
ultrasound, effectively reduces cell viability and induces apoptosis in the RASFs of RA patients35. Analysis of the genes that are epigenetically regulated in RA, taking into account not
only DNA methylation but also histone modifications, is worth exploring. In summary, we found that the proximal CpG motif −145C in the _IL10_ promoter region was hypomethylated in RA PBMCs,
which may regulate gene transcription and be responsible for the pathogenesis of RA. The demethylation of CpG motifs within the _IL10_ promoter contributes to its overexpression; thus,
demethylation may remedy the relative deficiency of IL10 in RA patients and may be a new therapeutic strategy in the treatment of RA. AUTHOR CONTRIBUTION Bin CONG designed the study; Li-hong
FU and Hai-ying CHEN performed the research; Chun-ling MA and Jing-ge ZHANG contributed new reagents or analytic tools; Li-hong FU and Shu-jin LI analyzed data; and Li-hong FU wrote the
paper. REFERENCES * Choy EH, Panayi GS . Cytokine pathways and joint inflammation in rheumatoid arthritis. _N Engl J Med_ 2001; 344: 907–16. Article CAS PubMed Google Scholar * Keystone
E, Wherry J, Grint P . IL-10 as a therapeutic strategy in the treatment of rheumatoid arthritis. _Rheum Dis Clin North Am_ 1998; 24: 629–39. Article CAS PubMed Google Scholar * Finnegan
A, Kaplan CD, Cao Y, Eibel H, Glant TT, Zhang J . Collagen-induced arthritis is exacerbated in IL-10-deficient mice. _Arthritis Res Ther_ 2003; 5: R18–24. Article CAS PubMed Google
Scholar * Johansson AC, Hansson AS, Nandakumar KS, Bäcklund J, Holmdahl R . IL-10-deficient B10.Q mice develop more severe collagen-induced arthritis, but are protected from arthritis
induced with anti-type II collagen antibodies. _J Immunol_ 2001; 167: 3505–12. Article CAS PubMed Google Scholar * Dai Q, Li Y, Zhang F, Yu H, Wang X . Therapeutic effect of low-dose
IL-18 combined with IL-10 on collagen-induced arthritis by down-regulation of inflammatory and Th1 responses and induction of Th2 responses. _Rheumatol Int_ 2009; 29: 615–22. Article CAS
PubMed Google Scholar * Choi JJ, Yoo SA, Park SJ, Kang YJ, Kim WU, Oh IH, _et al_. Mesenchymal stem cells overexpressing interleukin-10 attenuate collagen-induced arthritis in mice. _Clin
Exp Immunol_ 2008; 153: 269–76. Article CAS PubMed PubMed Central Google Scholar * Fitzpatrick DR, Wilson CB . Methylation and demethylation in the regulation of genes, cells, and
responses in the immune system. _Clin Immunol_ 2003; 109: 37–45. Article CAS PubMed Google Scholar * Dong J, Ivascu C, Chang HD, Wu P, Angeli R, Maggi L, _et al_. IL-10 is excluded from
the functional cytokine memory of human CD4+ memory T lymphocytes. _J Immunol_ 2007; 179: 2389–96. Article CAS PubMed Google Scholar * Tsuji-Takayama K, Suzuki M, Yamamoto M, Harashima
A, Okochi A, Otani T, _et al_. The production of IL-10 by human regulatory T cells is enhanced by IL-2 through a STAT5-responsive intronic enhancer in the IL-10 locus. _J Immunol_ 2008; 181:
3897–905. Article CAS PubMed Google Scholar * Gee K, Angel JB, Ma W, Mishra S, Gajanayaka N, Parato K, _et al_. Intracellular HIV-Tat expression induces IL-10 synthesis by the CREB-1
transcription factor through Ser133 phosphorylation and its regulation by the ERK1/2 MAPK in human monocytic cells. _J Biol Chem_ 2006; 281: 31647–58. Article CAS PubMed Google Scholar *
Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, _et al_. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. _Immunity_ 2006;
24: 563–74. Article CAS PubMed Google Scholar * Ishizu A, Abe A, Miyatake Y, Baba T, Iinuma C, Tomaru U, _et al_. Cyclic AMP response element-binding protein is implicated in IL-6
production from arthritic synovial cells. _Mod Rheumatol_ 2010; 20: 134–8. Article CAS PubMed Google Scholar * Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M .
Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. _Arthritis Rheum_ 1990; 33: 1665–73. Article CAS PubMed Google Scholar * Takami N,
Osawa K, Miura Y, Komai K, Taniguchi M, Shiraishi M, _et al_. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. _Arthritis Rheum_
2006; 54: 779–87. Article CAS PubMed Google Scholar * Nile CJ, Read RC, Akil M, Duff GW, Wilson AG . Methylation status of a single CpG site in the IL6 promoter is related to IL6
messenger RNA levels and rheumatoid arthritis. _Arthritis Rheum_ 2008; 58: 2686–93. Article PubMed Google Scholar * Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, _et
al_. The American rheumatoid association 1987 revised criteria for the classification of rheumatoid arthritis. _Arthritis Rheum_ 1988; 31: 315–24. Article CAS PubMed Google Scholar * Mi
XB, Zeng FQ . Hypomethylation of interleukin-4 and -6 promoters in T cells from systemic lupus erythematosus patients. _Acta Pharmacol Sin_ 2008; 29: 105–12. Article CAS PubMed Google
Scholar * Loots GG, Ovcharenko I . rVISTA 2.0: evolutionary analysis of transcription factor binding sites. _Nucleic Acids Res_ 2004; 32: W217–21. Article CAS PubMed PubMed Central
Google Scholar * Clark SJ, Harrison J, Paul CL, Frommer M . High sensitivity mapping of methylated cytosines. _Nucleic Acids Res_ 1994; 22: 2990–7. Article CAS PubMed PubMed Central
Google Scholar * van Roon JA, Bijlsma JW, Lafeber FP . Suppression of inflammation and joint destruction in rheumatoid arthritis may require a concerted action of Th2 cytokines. _Curr Opin
Investig Drugs_ 2002; 3: 1011–6. CAS PubMed Google Scholar * Whalen JD, Lechman EL, Carlos CA, Weiss K, Kovesdi I, Glorioso JC, _et al_. Adenoviral transfer of the viral IL10 gene
periarticularly to mouse paws suppresses development of collagen-induced arthritis in both injected and uninjected paws. _J Immunol_ 1999; 162: 3625–32. CAS PubMed Google Scholar * Im SH,
Hueber A, Monticelli S, Kang KH, Rao A . Chromatin-level regulation of the IL10 gene in T cells. _J Biol Chem_ 2004; 279: 46818–25. Article CAS PubMed Google Scholar * Iwata Y,
Matsushita T, Horikawa M, Dilillo DJ, Yanaba K, Venturi GM, _et al_. Characterization of a rare IL-10-competent B-cell subset in humans that parallels mouse regulatory B10 cells. _Blood_
2011; 117: 530–41. Article CAS PubMed PubMed Central Google Scholar * LIorente L, Richaud-Patin Y, Fior R, Alcocer-Varela J, Wijdenes J, Fourrier BM, _et al_. _In vivo_ production of
interleukin-10 by non-T cells in rheumatoid arthritis, Sjogren's syndrome, and systemic lupus erythematosus: a potential mechanism of B lymphocyte hyperactivity and autoimmunity.
_Arthritis Rheum_ 1994; 37: 1647–55. Article Google Scholar * Sempere-Ortells JM, Pérez-García V, Marín-Alberca G, Peris-Pertusa A, Benito JM, Marco FM, _et al_. Quantification and
phenotype of regulatory T cells in rheumatoid arthritis according to disease activity score-28. _Autoimmunity_ 2009; 42: 636–45. Article CAS PubMed Google Scholar * Xu J, Winkler J,
Sabarinath SN, Derendorf H . Assessment of the impact of dosing time on the pharmacokinetics/pharmacodynamics of prednisolone. _AAPS J_ 2008; 10: 331–41. Article CAS PubMed PubMed Central
Google Scholar * Archer TK, Deroo BJ, Fryer CJ . Chromatin modulation of glucocorticoid and progesterone receptor activity. _Trends Endocrinol Metab_ 1997; 8: 384–90. Article CAS PubMed
Google Scholar * Kim YI, Logan JW, Mason JB, Roubenoff R . DNA hypomethylation in inflammatory arthritis: reversal with methotrexate. _J Lab Clin Med_ 1996; 128: 165–72. Article CAS
PubMed Google Scholar * Issa JP, Vertino PM, Wu J, Sazawal S, Celano P, Nelkin BD, _et al_. Increased cytosine DNA-methyltransferase activity during colon cancer progression. _J Natl
Cancer Inst_ 1993; 85: 1235–40. Article CAS PubMed Google Scholar * Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA . Differential mRNA expression of the human DNA
methyltransferases (DNMTs) 1, 3a and 3b during the G0/G1 to S phase transition in normal and tumor cells. _Nucleic Acids Res_ 2000; 28: 2108–13. Article CAS PubMed PubMed Central Google
Scholar * Ramji DP, Foka P . CCAAT/enhancer-binding proteins: structure, function and regulation. _Biochem J_ 2002; 365: 561–75. Article CAS PubMed PubMed Central Google Scholar *
Benkhart EM, Siedlar M, Wedel A, Werner T, Ziegler-Heitbrock HW . Role of Stat3 in lipopolysaccharide-induced IL-10 gene expression. _J Immunol_ 2000; 165: 1612–7. Article CAS PubMed
Google Scholar * Huber LC, Brock M, Hemmatazad H, Giger OT, Moritz F, Trenkmann M, _et al_. Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis
and osteoarthritis patients. _Arthritis Rheum_ 2007; 56: 1087–93. Article CAS PubMed Google Scholar * Nishida K, Komiyama T, Miyazawa S, Shen ZN, Furumatsu T, Doi H, _et al_. Histone
deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21WAF1/Cip1 expression. _Arthritis Rheum_ 2004; 50: 3365–76. Article CAS PubMed
Google Scholar * Nakamura C, Matsushita I, Kosaka E, Kondo T, Kimura T . Anti-arthritic effects of combined treatment with histone deacetylase inhibitor and low-intensity ultrasound in
the presence of microbubbles in human rheumatoid synovial cells. _Rheumatology (Oxford)_ 2008; 47: 418–24. Article CAS Google Scholar Download references ACKNOWLEDGEMENTS Our sincere
thanks go to Mr Jian-gang WEI, PhD candidate of Shandong University, China, for his proofreading of the paper. Our work is supported by the National Natural Science Foundation of China
(grant 30770839) and the Hebei Province Natural Science Foundation of China (grant 30100154). We are grateful to the Second and Third Affiliated Hospitals of Hebei Medical University for
recruiting the RA patient samples. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Forensic Medicine, Hebei Key Laboratory of Forensic Medicine, Hebei Medical University,
Shijiazhuang, 050017, China Li-hong Fu, Chun-ling Ma, Bin Cong, Shu-jin Li, Hai-ying Chen & Jing-ge Zhang Authors * Li-hong Fu View author publications You can also search for this
author inPubMed Google Scholar * Chun-ling Ma View author publications You can also search for this author inPubMed Google Scholar * Bin Cong View author publications You can also search for
this author inPubMed Google Scholar * Shu-jin Li View author publications You can also search for this author inPubMed Google Scholar * Hai-ying Chen View author publications You can also
search for this author inPubMed Google Scholar * Jing-ge Zhang View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Bin
Cong. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Fu, Lh., Ma, Cl., Cong, B. _et al._ Hypomethylation of proximal CpG motif of interleukin-10
promoter regulates its expression in human rheumatoid arthritis. _Acta Pharmacol Sin_ 32, 1373–1380 (2011). https://doi.org/10.1038/aps.2011.98 Download citation * Received: 24 December 2010
* Accepted: 21 June 2011 * Published: 10 October 2011 * Issue Date: November 2011 * DOI: https://doi.org/10.1038/aps.2011.98 SHARE THIS ARTICLE Anyone you share the following link with will
be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative KEYWORDS * rheumatoid arthritis * interleukin-10 * DNA methylation * promoter regions * interspecies-conserved sequence * CpG motif * cyclic AMP response
element-binding protein * bioinformatics
