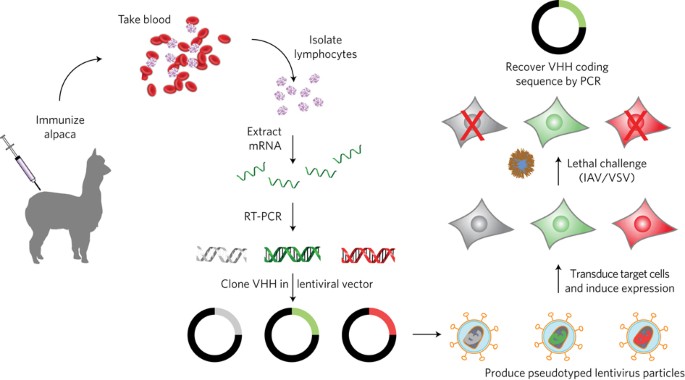
Phenotypic lentivirus screens to identify functional single domain antibodies
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Manipulation of proteins is key in assessing their _in vivo_ function. Although genetic ablation is straightforward, reversible and specific perturbation of protein function remains
a challenge. Single domain antibody fragments, such as camelid-derived VHHs, can serve as inhibitors or activators of intracellular protein function, but functional testing of identified
VHHs is laborious. To address this challenge, we have developed a lentiviral screening approach to identify VHHs that elicit a phenotype when expressed intracellularly. We identified 19
antiviral VHHs that protect human A549 cells from lethal infection with influenza A virus (IAV) or vesicular stomatitis virus (VSV), respectively. Both negative-sense RNA viruses are
vulnerable to VHHs uniquely specific for their respective nucleoproteins. Antiviral VHHs prevented nuclear import of viral ribonucleoproteins or mRNA transcription, respectively, and may
provide clues for novel antiviral reagents. In principle, the screening approach described here should be applicable to identify inhibitors of any pathogen or biological pathway. Access
through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal
Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS A VIRALLY ENCODED HIGH-RESOLUTION SCREEN OF CYTOMEGALOVIRUS DEPENDENCIES Article 05 June 2024 REPLICATION COMPETENT HIV-GUIDED CRISPR SCREEN
IDENTIFIES ANTIVIRAL FACTORS INCLUDING TARGETS OF THE ACCESSORY PROTEIN NEF Article Open access 07 May 2024 OMICRON ESCAPES THE MAJORITY OF EXISTING SARS-COV-2 NEUTRALIZING ANTIBODIES
Article Open access 23 December 2021 REFERENCES * Mohr, S. E., Smith, J. A., Shamu, C. E., Neumuller, R. A. & Perrimon, N. RNAi screening comes of age: improved techniques and
complementary approaches. _Nature Rev. Mol. Cell Biol._ 15, 591–600 (2014). Article Google Scholar * Kim, H. & Kim, J. S. A guide to genome engineering with programmable nucleases.
_Nature Rev. Genet._ 15, 321–334 (2014). Article Google Scholar * Cohen, P. Guidelines for the effective use of chemical inhibitors of protein function to understand their roles in cell
regulation. _Biochem. J._ 425, 53–54 (2010). Article Google Scholar * Doxsey, S. J., Brodsky, F. M., Blank, G. S. & Helenius, A. Inhibition of endocytosis by anti-clathrin antibodies.
_Cell_ 50, 453–463 (1987). Article Google Scholar * Gargano, N. & Cattaneo, A. Rescue of a neutralizing anti-viral antibody fragment from an intracellular polyclonal repertoire
expressed in mammalian cells. _FEBS Lett._ 414, 537–540 (1997). Article Google Scholar * Hamers-Casterman, C. _et al._ Naturally occurring antibodies devoid of light chains. _Nature_ 363,
446–448 (1993). Article Google Scholar * Muyldermans, S. Nanobodies: natural single-domain antibodies. _Annu. Rev. Biochem._ 82, 775–797 (2013). Article Google Scholar * Helma, J.,
Cardoso, M. C., Muyldermans, S. & Leonhardt, H. Nanobodies and recombinant binders in cell biology. _J. Cell Biol._ 209, 633–644 (2015). Article Google Scholar * Schmidt, F. I. _et
al._ A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. _J. Exp. Med._ 213, 771–790 (2016). Article Google Scholar
* Maass, D. R., Sepulveda, J., Pernthaner, A. & Shoemaker, C. B. Alpaca (_Lama pacos_) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). _J. Immunol. Methods_
324, 13–25 (2007). Article Google Scholar * Ryckaert, S., Pardon, E., Steyaert, J. & Callewaert, N. Isolation of antigen-binding camelid heavy chain antibody fragments (nanobodies)
from an immune library displayed on the surface of _Pichia pastoris_. _J. Biotechnol._ 145, 93–98 (2010). Article Google Scholar * Fridy, P. C. _et al._ A robust pipeline for rapid
production of versatile nanobody repertoires. _Nature Methods_ 11, 1253–1260 (2014). Article Google Scholar * Ashour, J. _et al._ Intracellular expression of camelid single-domain
antibodies specific for influenza virus nucleoprotein uncovers distinct features of its nuclear localization. _J. Virol._ 89, 2792–2800 (2015). Article Google Scholar * Dougan, S. K. _et
al._ Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. _Nature_ 503, 406–409 (2013). Article Google Scholar * Barrios-Rodiles, M. _et al._
High-throughput mapping of a dynamic signaling network in mammalian cells. _Science_ 307, 1621–1625 (2005). Article Google Scholar * Guimaraes, C. P. _et al._ Site-specific C-terminal and
internal loop labeling of proteins using sortase-mediated reactions. _Nature Protoc._ 8, 1787–1799 (2013). Article Google Scholar * Fodor, E. The RNA polymerase of influenza a virus:
mechanisms of viral transcription and replication. _Acta Virol._ 57, 113–122 (2013). Article Google Scholar * Lyles, D. S. & Rupprecht, C. E. in _Fields’ Virology_ (eds Fields, B. N.
_et al._ 1364–1408 (Wolters Kluwer Health/Lippincott Williams & Wilkins, 2007). Google Scholar * Emerson, S. U. & Wagner, R. R. Dissociation and reconstitution of the transcriptase
and template activities of vesicular stomatitis B and T virions. _J. Virol._ 10, 297–309 (1972). Google Scholar * Green, T. J. & Luo, M. Structure of the vesicular stomatitis virus
nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. _Proc. Natl Acad. Sci. USA_ 106, 11713–11718 (2009). Article Google Scholar * Rahmeh, A.
A. _et al._ Molecular architecture of the vesicular stomatitis virus RNA polymerase. _Proc. Natl Acad. Sci. USA_ 107, 20075–20080 (2010). Article Google Scholar * Baltimore, D., Huang, A.
S. & Stampfer, M. Ribonucleic acid synthesis of vesicular stomatitis virus, II. An RNA polymerase in the virion. _Proc. Natl Acad. Sci. USA_ 66, 572–576 (1970). Article Google Scholar
* Li, T. _et al._ Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood–brain barrier and specifically recognize astrocytes:
application to brain imaging. _FASEB J._ 26, 3969–3979 (2012). Article Google Scholar * Morin, B., Kranzusch, P. J., Rahmeh, A. A. & Whelan, S. P. The polymerase of negative-stranded
RNA viruses. _Curr. Opin. Virol._ 3, 103–110 (2013). Article Google Scholar * Yewdell, J. W., Frank, E. & Gerhard, W. Expression of influenza A virus internal antigens on the surface
of infected P815 cells. _J. Immunol._ 126, 1814–1819 (1981). Google Scholar * Sosa, B. A. _et al._ How lamina-associated polypeptide 1 (LAP1) activates torsin. _eLife_ 3, e03239 (2014).
Article Google Scholar * Meerbrey, K. L. _et al._ The pINDUCER lentiviral toolkit for inducible RNA interference _in vitro_ and _in vivo_. _Proc. Natl Acad. Sci. USA_ 108, 3665–3670
(2011). Article Google Scholar * Lefrancois, L. & Lyles, D. S. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of
neutralizing epitopes with monoclonal antibodies. _Virology_ 121, 157–167 (1982). Article Google Scholar * Taipale, M. _et al._ Quantitative analysis of HSP90–client interactions reveals
principles of substrate recognition. _Cell_ 150, 987–1001 (2012). Article Google Scholar * Conrath, K. E. _et al._ Beta-lactamase inhibitors derived from single-domain antibody fragments
elicited in the camelidae. _Antimicrob. Agents Chemother._ 45, 2807–2812 (2001). Article Google Scholar * Andersen, K. R., Leksa, N. C. & Schwartz, T. U. Optimized _E. coli_ expression
strain LOBSTR eliminates common contaminants from His-tag purification. _Proteins_ 81, 1857–1861 (2013). Article Google Scholar * Green, T. J. _et al._ Access to RNA encapsidated in the
nucleocapsid of vesicular stomatitis virus. _J. Virol._ 85, 2714–2722 (2011). Article Google Scholar * Kamentsky, L. _et al._ Improved structure, function and compatibility for
CellProfiler: modular high-throughput image analysis software. _Bioinformatics_ 27, 1179–1180 (2011). Article Google Scholar * Schickli, J. H. _et al._ Plasmid-only rescue of influenza A
virus vaccine candidates. _Phil Trans. R. Soc. Lond. B_ 356, 1965–1973 (2001). Article Google Scholar * Fodor, E. _et al._ Rescue of influenza A virus from recombinant DNA. _J. Virol._ 73,
9679–9682 (1999). Google Scholar * Pattnaik, A. K. & Wertz, G. W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing
viral proteins from vectors containing cloned cDNAs. _J. Virol._ 64, 2948–2957 (1990). Google Scholar * Lehrach, H., Diamond, D., Wozney, J. M. & Boedtker, H. RNA molecular weight
determinations by gel electrophoresis under denaturing conditions, a critical reexamination. _Biochemistry_ 16, 4743–4751 (1977). Article Google Scholar * Morin, B., Rahmeh, A. A. &
Whelan, S. P. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. _EMBO J._ 31, 1320–1329 (2012). Article Google Scholar * Cherry, S. _et al._ Genome-wide
RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. _Genes Dev._ 19, 445–452 (2005). Article Google Scholar Download references
ACKNOWLEDGEMENTS The authors thank B. Bierie for help with lentiviral vectors, M. Taipale and G. Karras for help with LUMIER assays, S. Hulsey Stubbs for VSV-neutralizing antibodies, and T.
DiCesare for help with illustrations. This work is supported by a National Institutes of Health Pioneer award to H.L.P. and additional funding from Fujifilm/MediVector. F.I.S. was supported
by an Advanced Postdoc.Mobility Fellowship from the Swiss National Science Foundation (SNSF). AUTHOR INFORMATION Author notes * Benjamin Morin Present address: † Present address: Agenus
Inc., Lexington, Massachusetts 02421, USA., AUTHORS AND AFFILIATIONS * Whitehead Institute for Biomedical Research, Cambridge, 02142, Massachusetts, USA Florian I. Schmidt, Leo Hanke,
Rebeccah Brewer & Hidde L. Ploegh * Department of Microbiology and Immunobiology, Harvard Medical School, Boston, 02115, Massachusetts, USA Benjamin Morin, Vesna Brusic & Sean P.J.
Whelan * Department of Biology, Massachusetts Institute of Technology, Cambridge, 02139, Massachusetts, USA Hidde L. Ploegh Authors * Florian I. Schmidt View author publications You can also
search for this author inPubMed Google Scholar * Leo Hanke View author publications You can also search for this author inPubMed Google Scholar * Benjamin Morin View author publications You
can also search for this author inPubMed Google Scholar * Rebeccah Brewer View author publications You can also search for this author inPubMed Google Scholar * Vesna Brusic View author
publications You can also search for this author inPubMed Google Scholar * Sean P.J. Whelan View author publications You can also search for this author inPubMed Google Scholar * Hidde L.
Ploegh View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS F.I.S., L.H., B.M., R.B. and V.B. performed experiments and analysed the data.
S.P.J.W. gave critical technical advice. F.I.S. and H.L.P. conceived the study and wrote the manuscript. CORRESPONDING AUTHOR Correspondence to Hidde L. Ploegh. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figures 1-7 (PDF 866 kb) RIGHTS AND PERMISSIONS Reprints
and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Schmidt, F., Hanke, L., Morin, B. _et al._ Phenotypic lentivirus screens to identify functional single domain antibodies. _Nat Microbiol_
1, 16080 (2016). https://doi.org/10.1038/nmicrobiol.2016.80 Download citation * Received: 28 January 2016 * Accepted: 28 April 2016 * Published: 20 June 2016 * DOI:
https://doi.org/10.1038/nmicrobiol.2016.80 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
