Lipid bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state nmr samples
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

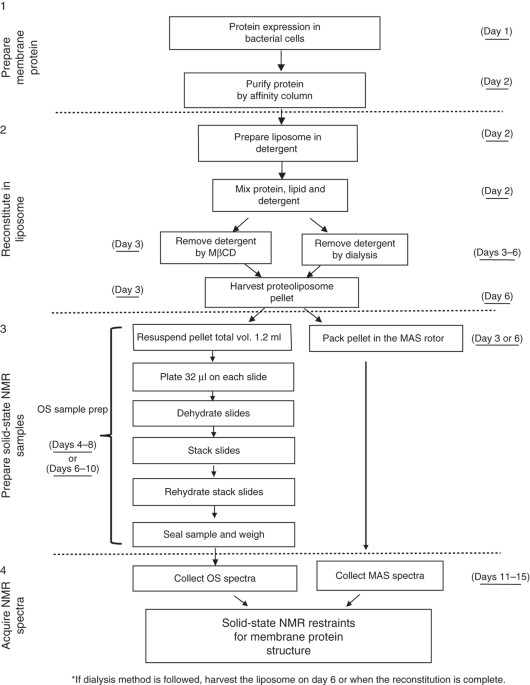
ABSTRACT Solid-state NMR spectroscopy has been used successfully for characterizing the structure and dynamics of membrane proteins as well as their interactions with other proteins in lipid
bilayers. Such an environment is often necessary for achieving native-like structures. Sample preparation is the key to this success. Here we present a detailed description of a robust
protocol that results in high-quality membrane protein samples for both magic-angle spinning and oriented-sample solid-state NMR. The procedure is demonstrated using two proteins: CrgA (two
transmembrane helices) and Rv1861 (three transmembrane helices), both from _Mycobacterium tuberculosis_. The success of this procedure relies on two points. First, for samples for both types
of NMR experiment, the reconstitution of the protein from a detergent environment to an environment in which it is incorporated into liposomes results in 'complete' removal of
detergent. Second, for the oriented samples, proper dehydration followed by rehydration of the proteoliposomes is essential. By using this protocol, proteoliposome samples for magic-angle
spinning NMR and uniformly aligned samples (orientational mosaicity of <1°) for oriented-sample NMR can be obtained within 10 d. Access through your institution Buy or subscribe This is a
preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per
year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS A ‘BUILD AND
RETRIEVE’ METHODOLOGY TO SIMULTANEOUSLY SOLVE CRYO-EM STRUCTURES OF MEMBRANE PROTEINS Article 06 January 2021 THE IMPORTANCE OF THE MEMBRANE FOR BIOPHYSICAL MEASUREMENTS Article 16 November
2020 STABILIZATION AND STRUCTURE DETERMINATION OF INTEGRAL MEMBRANE PROTEINS BY TERMINI RESTRAINING Article 17 January 2022 REFERENCES * Page, R.C., Li, C., Hu, J., Gao, F.P. & Cross,
T.A. Lipid bilayers: an essential environment for the understanding of membrane proteins. _Magn. Reson. Chem._ 45, S2–S11 (2007). Article CAS Google Scholar * Dong, H., Sharma, M., Zhou,
H.X. & Cross, T.A. Glycines: role in α-helical membrane protein structures and a potential indicator of native conformation. _Biochemistry_ 51, 4779–4789 (2012). Article CAS Google
Scholar * Zhou, H.X. & Cross, T.A. Influences of membrane mimetic environments on membrane protein structures. _Annu. Rev. Biophys._ 42, 361–392 (2013). Article CAS Google Scholar *
Anfinsen, C.B. Principles that govern the folding of protein chains. _Science_ 181, 223–230 (1973). Article CAS Google Scholar * Korepanova, A. et al. Cloning and expression of multiple
integral membrane proteins from _Mycobacterium tuberculosis_ in _Escherichia coli_. _Protein Sci._ 14, 148–158 (2005). Article CAS Google Scholar * Caffrey, M. Crystallizing membrane
proteins for structure determination: use of lipidic mesophases. _Annu. Rev. Biophys._ 38, 29–51 (2009). Article CAS Google Scholar * Separovic, F., Killian, J.A., Cotten, M., Busath,
D.D. & Cross, T.A. Modeling the membrane environment for membrane proteins. _Biophys. J._ 100, 2073–2074 (2011). Article CAS Google Scholar * Li, D. et al. Crystal structure of the
integral membrane diacylglycerol kinase. _Nature_ 497, 521–524 (2013). Article CAS Google Scholar * Chou, J.J., Kaufman, J.D., Stahl, S.J., Wingfield, P.T. & Bax, A. Micelle-induced
curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. _J. Am. Chem. Soc._ 124, 2450–2451 (2002). Article CAS Google
Scholar * Van Horn, W.D. et al. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. _Science_ 324, 1726–1729 (2009). Article CAS Google Scholar *
Griffin, R.G. Dipolar recoupling in MAS spectra of biological solids. _Nat. Struct. Biol._ 5, 508–512 (1998). Article CAS Google Scholar * Tang, W., Knox, R.W. & Nevzorov, A.A. A
spectroscopic assignment technique for membrane proteins reconstituted in magnetically aligned bicelles. _J. Biomol. NMR_ 54, 307–316 (2012). Article CAS Google Scholar * Ketchem, R.R.,
Hu, W., Tian, F. & Cross, T.A. Structure and dynamics from solid state NMR spectroscopy. _Structure_ 2, 699–701 (1994). Article CAS Google Scholar * Sharma, M. et al. Insight into the
mechanism of the influenza A proton channel from a structure in a lipid bilayer. _Science_ 330, 509–512 (2010). Article CAS Google Scholar * Mote, K.R. et al. Multidimensional oriented
solid-state NMR experiments enable the sequential assignment of uniformly 15N labeled integral membrane proteins in magnetically aligned lipid bilayers. _Journal of Biomolecular NMR_ 51,
339–346 (2011). Article CAS Google Scholar * Nevzorov, A.A. & Opella, S.J. Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. _J. Magn. Reson._
185, 59–70 (2007). Article CAS Google Scholar * Can, T.V. et al. Magic angle spinning and oriented sample solid-state NMR structural restraints combine for influenza A M2 protein
functional insights. _J. Am. Chem. Soc._ 134, 9022–9025 (2012). Article CAS Google Scholar * Murray, D.T., Das, N. & Cross, T.A. Solid-state NMR strategy for characterizing native
membrane protein structures. _Accounts Chem. Res._ 46, 2172–2181 (2013). Article CAS Google Scholar * Plocinski, P. et al. _Mycobacterium tuberculosis_ CwsA interacts with CrgA and Wag31,
and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. _J. Bacteriol._ 194, 6398–6409 (2012). Article CAS Google Scholar * Plocinski, P. et al.
Characterization of CrgA, a new partner of the _Mycobacterium tuberculosis_ peptidoglycan polymerization complexes. _J. Bacteriol._ 193, 3246–3256 (2011). Article CAS Google Scholar * Li,
C. et al. Uniformly aligned full-length membrane proteins in liquid crystalline bilayers for structural characterization. _J. Am. Chem. Soc._ 129, 5304–5305 (2007). Article CAS Google
Scholar * Su, P.C., Si, W., Baker, D.L. & Berger, B.W. High-yield membrane protein expression from _E. coli_ using an engineered outer membrane protein F fusion. _Protein Sci._ 22,
434–443 (2013). Article CAS Google Scholar * Grisshammer, R. Understanding recombinant expression of membrane proteins. _Curr. Opin. Biotechnol._ 17, 337–340 (2006). Article CAS Google
Scholar * Zoonens, M. & Miroux, B. Expression of membrane proteins at the _Escherichia coli_ membrane for structural studies. _Methods Mol. Biol._ 601, 49–66 (2010). Article CAS
Google Scholar * Hu, J., Qin, H., Gao, F.P. & Cross, T.A. A systematic assessment of mature MBP in membrane protein production: overexpression, membrane targeting and purification.
_Protein Expr. Purif._ 80, 34–40 (2011). Article CAS Google Scholar * Mouillac, B. & Baneres, J.L. Mammalian membrane receptors expression as inclusion bodies in _Escherichia coli_.
_Methods Mol. Biol._ 601, 39–48 (2010). Article CAS Google Scholar * Qin, H. et al. Construction of a series of vectors for high throughput cloning and expression screening of membrane
proteins from _Mycobacterium tuberculosis_. _BMC Biotechnol._ 8, 51 (2008). Article Google Scholar * Kiefer, H. et al. Expression of an olfactory receptor in _Escherichia coli_:
purification, reconstitution, and ligand binding. _Biochemistry_ 35, 16077–16084 (1996). Article CAS Google Scholar * Aslanidis, C. & de Jong, P.J. Ligation-independent cloning of PCR
products (LIC-PCR). _Nucleic Acids Res._ 18, 6069–6074 (1990). Article CAS Google Scholar * Korepanova, A. et al. Expression of membrane proteins from _Mycobacterium tuberculosis_ in
_Escherichia coli_ as fusions with maltose binding protein. _Protein Expr. Purif._ 53, 24–30 (2007). Article CAS Google Scholar * Seddon, A.M., Curnow, P. & Booth, P.J. Membrane
proteins, lipids and detergents: not just a soap opera. _Biochim. Biophys. Acta_ 1666, 105–117 (2004). Article CAS Google Scholar * Rigaud, J.L., Pitard, B. & Levy, D. Reconstitution
of membrane proteins into liposomes: application to energy-transducing membrane proteins. _Biochim. Biophys. Acta_ 1231, 223–246 (1995). Article Google Scholar * Rigaud, J.L. & Levy,
D. Reconstitution of membrane proteins into liposomes. _Methods Enzymol._ 372, 65–86 (2003). Article CAS Google Scholar * Signorell, G.A., Kaufmann, T.C., Kukulski, W., Engel, A. &
Remigy, H.W. Controlled 2D crystallization of membrane proteins using methyl-beta-cyclodextrin. _J. Struct. Biol._ 157, 321–328 (2007). Article CAS Google Scholar * DeGrip, W.J.,
VanOostrum, J. & Bovee-Geurts, P.H.M. Selective detergent-extraction from mixed detergent/lipid/protein micelles, using cyclodextrin inclusion compounds: a novel generic approach for the
preparation of proteoliposomes. _Biochem. J._ 330, 667–674 (1998). Article CAS Google Scholar * Rigaud, J.L., Levy, D., Mosser, G. & Lambert, O. Detergent removal by non-polar
polystyrene beads—applications to membrane protein reconstitution and two-dimensional crystallization. _Eur. Biophy. J. Biophys. Lett._ 27, 305–319 (1998). Article CAS Google Scholar *
Rigaud, J.L., Mosser, G., Lacapere, J.J., Olofsson, A., Levy, D. & Ranck, J.L. Bio-Beads: an efficient strategy for two-dimensional crystallization of membrane proteins. _J. Struct.
Biol._ 118, 226–235 (1997). Article CAS Google Scholar * Kimura, T. et al. Recombinant cannabinoid type 2 receptor in liposome model activates G protein in response to anionic lipid
constituents. _J. Biol. Chem._ 287, 4076–4087 (2012). Article CAS Google Scholar * Park, S.H. et al. Optimization of purification and refolding of the human chemokine receptor CXCR1
improves the stability of proteoliposomes for structure determination. _Biochim. Biophys. Acta_ 1818, 584–591 (2012). Article CAS Google Scholar * van den Brink-van der Laan, E., Killian,
J.A. & de Kruijff, B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. _Biochim. Biophys. Acta_ 1666, 275–288 (2004).
Article CAS Google Scholar * Miao, Y.M. et al. M2 proton channel structural validation from full-length protein samples in synthetic bilayers and _E. coli_ membranes. _Angew. Chem. Int.
Ed. Engl._ 51, 8383–8386 (2012). Article CAS Google Scholar * Cady, S., Wang, T. & Hong, M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the
influenza virus M2 protein. _J. Am. Chem. Soc._ 133, 11572–11579 (2011). Article CAS Google Scholar * Bhate, M.P., Wylie, B.J., Tian, L. & McDermott, A.E. Conformational dynamics in
the selectivity filter of KcsA in response to potassium ion concentration. _J. Mol. Biol._ 401, 155–166 (2010). Article CAS Google Scholar * Peterson, E. et al. Functional reconstitution
of influenza A M2(22-62). _Biochim. Biophys. Acta_ 1808, 516–521 (2011). Article CAS Google Scholar * Nagy, J.K. & Sanders, C.R. Destabilizing mutations promote membrane protein
misfolding. _Biochemistry_ 43, 19–25 (2004). Article CAS Google Scholar * Wallace, B.A., Lees, J.G., Orry, A.J., Lobley, A. & Janes, R.W. Analyses of circular dichroism spectra of
membrane proteins. _Protein Sci._ 12, 875–884 (2003). Article CAS Google Scholar * Andreas, L.B., Eddy, M.T., Pielak, R.M., Chou, J. & Griffin, R.G. Magic angle spinning NMR
investigation of influenza A M2(18-60): support for an allosteric mechanism of inhibition. _J. Am. Chem Soc._ 132, 10958–10960 (2010). Article CAS Google Scholar * Ketchem, R.R., Hu, W.
& Cross, T.A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. _Science_ 261, 1457–1460 (1993). Article CAS Google Scholar * De Angelis, A.A. &
Opella, S.J. Bicelle samples for solid-state NMR of membrane proteins. _Nat. Protoc._ 2, 2332–2338 (2007). Article CAS Google Scholar * Cevc, G. & Marsh, D. _Phospholipid Bilayers:
Physical Principles and Models_ Vol. 5 (John Wiley & Sons, 1987). * Baldus, M. Solid-state nuclear magnetic resonance. In _Comprehensive Biophysics Vol. 1_ (ed. Engleman, E.) 160–181
(Academic Press, 2012). * Lakatos, A., Mors, K. & Glaubitz, C. How to investigate interactions between membrane proteins and ligands by solid-state NMR. _Methods Mol. Biol._ 914, 65–86
(2012). CAS PubMed Google Scholar * Wu, C.H., Ramamoorthy, A. & Opella, S.J. High resolution heteronuclear dipolar solid-state NMR spectroscopy. _J. Magn. Reson. A_ 109, 270–272
(1994). Article Google Scholar * Wang, J. et al. Imaging membrane protein helical wheels. _J. Magn. Reson._ 144, 162–167 (2000). Article CAS Google Scholar * Marassi, F.M. & Opella,
S.J.A solid-state NMR index of helical membrane protein structure and topology. _J. Magn. Reson._ 144, 150–155 (2000). Article CAS Google Scholar * Murray, D.T., Lu, Y.T., Cross, T.A.
& Quine, J.R. Geometry of kinked protein helices from NMR data. _J. Magn. Reson._ 210, 82–89 (2011). Article CAS Google Scholar * Opella, S.J. et al. Structures of the M2
channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. _Nat. Struct. Biol._ 6, 374–379 (1999). Article CAS Google Scholar * Park, S.H., De Angelis,
A.A., Nevzorov, A.A., Wu, C.H. & Opella, S.J. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. _Biophys. J._ 91, 3032–3042
(2006). Article CAS Google Scholar * Verardi, R., Shi, L., Traaseth, N.J., Walsh, N. & Veglia, G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution
and solid-state NMR method. _Proc. Natl. Acad. Sci. USA_ 108, 9101–9106 (2011). Article CAS Google Scholar * Zech, S.G., Wand, A.J. & McDermott, A.E. Protein structure determination
by high-resolution solid-state NMR spectroscopy: application to microcrystalline ubiquitin. _J. Am. Chem. Soc._ 127, 8618–8626 (2005). Article CAS Google Scholar * McDermott, A. Structure
and dynamics of membrane proteins by magic angle spinning solid-state NMR. _Annu. Rev. Biophys._ 38, 385–403 (2009). Article CAS Google Scholar * Castellani, F. et al. Structure of a
protein determined by solid-state magic-angle-spinning NMR spectroscopy. _Nature_ 420, 98–102 (2002). Article CAS Google Scholar * Park, S.H. et al. Structure of the chemokine receptor
CXCR1 in phospholipid bilayers. _Nature_ 491, 779–783 (2012). Article CAS Google Scholar * Das, B.B. et al. Structure determination of a membrane protein in proteoliposomes. _J. Am. Chem.
Soc._ 134, 2047–2056 (2012). Article CAS Google Scholar * Park, S.H., Das, B.B., De Angelis, A.A., Scrima, M. & Opella, S.J. Mechanically, magnetically, and 'rotationally
aligned' membrane proteins in phospholipid bilayers give equivalent angular constraints for NMR structure determination. _J. Phys. Chem. B_ 114, 13995–14003 (2010). Article CAS Google
Scholar * Opella, S.J. Structure determination of membrane proteins in their native phospholipid bilayer environment by rotationally aligned solid-state NMR spectroscopy. _Accounts Chem.
Res._ 46, 2145–2153 (2013). Article CAS Google Scholar * Cady, S.D. et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. _Nature_ 463,
689–692 (2010). Article CAS Google Scholar * Massotte, D. G protein-coupled receptor overexpression with the baculovirus-insect cell system: a tool for structural and functional studies.
_Biochim. Biophys. Acta_ 1610, 77–89 (2003). Article CAS Google Scholar * McCusker, E.C., Bane, S.E., O'Malley, M.A. & Robinson, A.S. Heterologous GPCR expression: a bottleneck
to obtaining crystal structures. _Biotechnol. Prog._ 23, 540–547 (2007). Article CAS Google Scholar * Shi, L. & Ladizhansky, V. Magic-angle spinning solid-state NMR experiments for
structural characterization of proteins. _Methods Mol. Biol._ 895, 153–165 (2012). Article CAS Google Scholar * Gor'kov, P.L. et al. Using low-E resonators to reduce RF heating in
biological samples for static solid-state NMR up to 900 MHz. _J. Magn. Reson._ 185, 77–93 (2007). Article CAS Google Scholar * Quine, J.R. et al. Intensity and mosaic spread analysis from
PISEMA tensors in solid-state NMR. _J. Magn. Reson._ 179, 190–198 (2006). Article CAS Google Scholar * Page, R.C. et al. Comprehensive evaluation of solution nuclear magnetic resonance
spectroscopy sample preparation for helical integral membrane proteins. _J. Struct. Funct. Genomics_ 7, 51–64 (2006). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We
thank M.W. Davidson at NHMFL and C. Escobar at FSU, IMB, NHMFL for helping with photography. We also thank P.L. Gor'kov for his design of the OS sample holder and the sample transfer
base. This work was supported in part by the US National Institutes of Health (grants AI 074805, AI 073891 and AI 023007) and the US National Science Foundation (through Cooperative
Agreement 0654118 between the Division of Materials Research and the State of Florida). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Institute of Molecular Biophysics (IMB), Florida State
University (FSU), Tallahassee, Florida, USA Nabanita Das, Dylan T Murray & Timothy A Cross * National High Magnetic Field Laboratory (NMHFL), FSU, Tallahassee, Florida, USA Nabanita Das,
Dylan T Murray & Timothy A Cross * Department of Chemistry and Biochemistry, FSU, Tallahassee, Florida, USA Timothy A Cross Authors * Nabanita Das View author publications You can also
search for this author inPubMed Google Scholar * Dylan T Murray View author publications You can also search for this author inPubMed Google Scholar * Timothy A Cross View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS N.D. and D.T.M. performed all the experiments such as membrane protein expression, purification,
solid-state NMR sample preparation and new method development. N.D. prepared all the figures. N.D. and T.A.C. wrote the manuscript, D.T.M. provided essential comments. All three authors
coordinated to complete this manuscript. CORRESPONDING AUTHOR Correspondence to Timothy A Cross. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests.
INTEGRATED SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 12% (WT/VOL) SDS-PAGE OF CRGA AND RV1861 MEMBRANE PROTEIN EXPRESSION AND PURIFICATION STEPS. M: Molecular weight marker, L: Whole
cell lysate containing inclusion body and membrane fractions, FT: Flow through from nickel column, Washes: two to three consecutive washing steps, Elutions: Protein elution from nickel
column. Molecular weights of the proteins are shown by red color arrows. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 12% (wt/vol) SDS-PAGE of CrgA and Rv1861 membrane protein expression
and purification steps. (PDF 3068 kb) SUPPLEMENTARY METHODS CrgA and Rv1861 membrane protein expression and purification; Reconstitution and OS sample preparation of 15N uniform labeled
gramicidin A protein in DMPC lipid bilayers. (PDF 208 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Das, N., Murray, D. & Cross, T. Lipid
bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state NMR samples. _Nat Protoc_ 8, 2256–2270 (2013). https://doi.org/10.1038/nprot.2013.129 Download
citation * Published: 24 October 2013 * Issue Date: November 2013 * DOI: https://doi.org/10.1038/nprot.2013.129 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative
