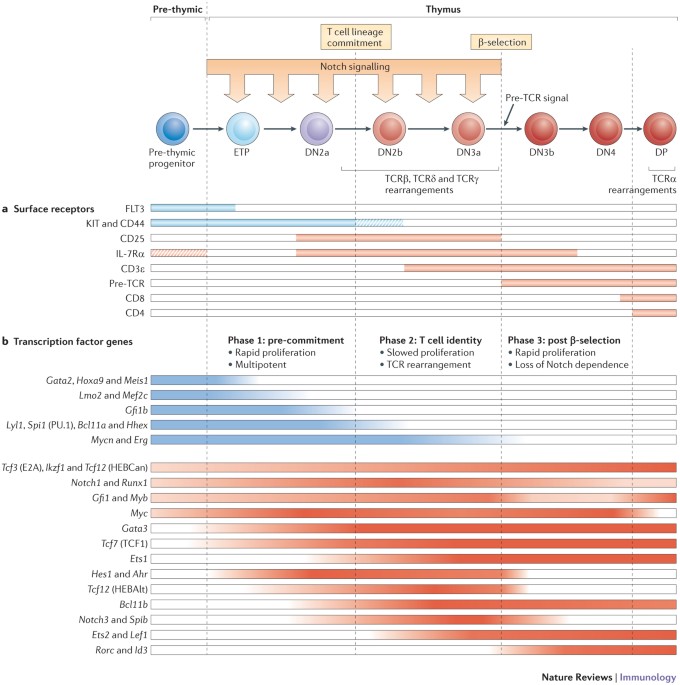
Developmental gene networks: a triathlon on the course to t cell identity
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

KEY POINTS * T cell development depends on the regulated progression of progenitor cells through three major phases that are associated with distinct transcription factor ensembles and
altered gene regulatory network states. * The first two phases are Notch dependent and T cell receptor (TCR) independent, whereas the third phase is TCR dependent and mediates the transition
to Notch independence. * The first phase is dominated by legacy stem and progenitor cell gene regulatory networks that enable self-renewal and Notch-dependent expression of the first T
cell-specific transcription factors, including GATA-binding protein 3 (GATA3) and T cell factor 1 (TCF1). * T cell lineage commitment — the loss of alternative latent developmental
potentials — is triggered by induction of the phase 2 transcription factor B cell lymphoma–leukaemia 11B (BCL11B) and results in the downregulation of multiple progenitor-specific
transcription factors. * In phase 2, T cell-specific regulators and Notch signalling drive the full activation of the T cell gene regulatory programme. TCR complex signalling components are
expressed, TCR gene recombination is induced and TCR-dependent selection thresholds are imposed. * Transition from phase 2 to phase 3 depends on the expression and successful signalling of a
pre-TCR or γδTCR. If successful, this switch from the phase 2 to phase 3 network triggers proliferation, loss of Notch dependency and dismantling of the Notch-dependent gene network. *
Phase 1 legacy and stem cell transcription factors can become oncogenic if their expression is not correctly controlled. Cells that fail to fully shut off the expression of phase 1
regulators at the commitment and β-selection checkpoints — before the next phase of gene network expression is activated — may be predisposed to leukaemic transformation. ABSTRACT Cells
acquire their ultimate identities by activating combinations of transcription factors that initiate and sustain expression of the appropriate cell type-specific genes. T cell development
depends on the progression of progenitor cells through three major phases, each of which is associated with distinct transcription factor ensembles that control the recruitment of these
cells to the thymus, their proliferation, lineage commitment and responsiveness to T cell receptor signals, all before the allocation of cells to particular effector programmes. All three
phases are essential for proper T cell development, as are the mechanisms that determine the boundaries between each phase. Cells that fail to shut off one set of regulators before the next
gene network phase is activated are predisposed to leukaemic transformation. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this
article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in
* Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS RUNX FACTORS LAUNCH T CELL AND INNATE LYMPHOID PROGRAMS VIA DIRECT
AND GENE NETWORK-BASED MECHANISMS Article 10 August 2023 HOW TRANSCRIPTION FACTORS DRIVE CHOICE OF THE T CELL FATE Article 11 September 2020 T-CELL COMMITMENT INHERITANCE—AN AGENT-BASED
MULTI-SCALE MODEL Article Open access 17 April 2024 REFERENCES * Rothenberg, E. V., Moore, J. E. & Yui, M. A. Launching the T-cell-lineage developmental programme. _Nature Rev. Immunol._
8, 9–21 (2008). CAS Google Scholar * Petrie, H. T. & Zúñiga-Pflücker, J. C. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. _Annu. Rev. Immunol._
25, 649–679 (2007). CAS PubMed Google Scholar * Thompson, P. K. & Zúñiga-Pflücker, J. C. On becoming a T cell, a convergence of factors kick it up a Notch along the way. _Semin.
Immunol._ 23, 350–359 (2011). CAS PubMed Google Scholar * Yang, Q., Jeremiah Bell, J. J. & Bhandoola, A. T-cell lineage determination. _Immunol. Rev._ 238, 12–22 (2010). CAS PubMed
PubMed Central Google Scholar * Koch, U. & Radtke, F. Mechanisms of T cell development and transformation. _Annu. Rev. Cell Dev. Biol._ 27, 539–562 (2011). CAS PubMed Google Scholar
* Love, P. E. & Bhandoola, A. Signal integration and crosstalk during thymocyte migration and emigration. _Nature Rev. Immunol._ 11, 469–477 (2011). CAS Google Scholar * Lu, M. et
al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. _J. Immunol._
175, 5848–5856 (2005). CAS PubMed Google Scholar * Porritt, H. E., Gordon, K. & Petrie, H. T. Kinetics of steady-state differentiation and mapping of intrathymic-signaling
environments by stem cell transplantation in nonirradiated mice. _J. Exp. Med._ 198, 957–962 (2003). CAS PubMed PubMed Central Google Scholar * Belyaev, N. N., Biro, J., Athanasakis, D.,
Fernandez-Reyes, D. & A. J. Global transcriptional analysis of primitive thymocytes reveals accelerated dynamics of T cell specification in fetal stages. _Immunogenetics_ 64, 591–604
(2012). CAS PubMed PubMed Central Google Scholar * Schmitt, T. M. & Zúñiga-Pflücker, J. C. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 _in
vitro_. _Immunity_ 17, 749–756 (2002). THIS PAPER TRANSFORMED THE FIELD OF EARLY T CELL DEVELOPMENT BY CREATING A POWERFUL AND EFFICIENT _IN VITRO_ SYSTEM IN WHICH THE DEVELOPMENTAL PROCESS
CAN BE OBSERVED AND MANIPULATED. CAS PubMed Google Scholar * Ng, S. Y., Yoshida, T., Zhang, J. & Georgopoulos, K. Genome-wide lineage-specific transcriptional networks underscore
Ikaros-dependent lymphoid priming in hematopoietic stem cells. _Immunity_ 30, 493–507 (2009). CAS PubMed PubMed Central Google Scholar * Chi, A. W. et al. Identification of Flt3+CD150−
myeloid progenitors in adult mouse bone marrow that harbor T lymphoid developmental potential. _Blood_ 118, 2723–2732 (2011). CAS PubMed PubMed Central Google Scholar * Krueger, A. &
von Boehmer, H. Identification of a T lineage-committed progenitor in adult blood. _Immunity_ 26, 105–116 (2007). CAS PubMed PubMed Central Google Scholar * Boudil, A. et al.
Single-cell analysis of thymocyte differentiation: identification of transcription factor interactions and a major stochastic component in αβ-lineage commitment. _PLoS ONE_ 8, e73098 (2013).
CAS PubMed PubMed Central Google Scholar * Peaudecerf, L. et al. Thymocytes may persist and differentiate without any input from bone marrow progenitors. _J. Exp. Med._ 209, 1401–1408
(2012). CAS PubMed PubMed Central Google Scholar * Martins, V. C. et al. Thymus-autonomous T cell development in the absence of progenitor import. _J. Exp. Med._ 209, 1409–1417 (2012).
CAS PubMed PubMed Central Google Scholar * Martins, V. C. et al. Cell competition is a tumour suppressor mechanism in the thymus. _Nature_ 509, 465–470 (2014). REFERENCES 15–17
OVERTURNED THE LONG-STANDING THEORY THAT THYMOCYTES CANNOT SELF-RENEW. THEY SHOW THAT THE LOSS OF COMPETITION FROM FRESH THYMIC IMMIGRANTS LEADS TO EXTENSIVE SELF-RENEWAL AND ALLOWS
ONCOGENIC TRANSFORMATION OF EARLY STAGE THYMOCYTES. CAS PubMed Google Scholar * Coustan-Smith, E. et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic
leukaemia. _Lancet Oncol._ 10, 147–156 (2009). CAS PubMed PubMed Central Google Scholar * Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia.
_Nature_ 481, 157–163 (2012). REFERENCES 18 AND 19 CHARACTERIZE ETP-ALL, A HIGH-RISK HUMAN T-ALL SUBTYPE THAT IS CHARACTERIZED BY THE EXPRESSION OF GENES ASSOCIATED WITH THE VERY EARLIEST
STAGES OF NORMAL T CELL DEVELOPMENT. CAS PubMed PubMed Central Google Scholar * Schlenner, S. M. et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the
thymus. _Immunity_ 32, 426–436 (2010). CAS PubMed Google Scholar * Luc, S. et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. _Nature Immunol._
13, 412–419 (2012). CAS Google Scholar * Serwold, T., Ehrlich, L. I. & Weissman, I. L. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the
major source of thymopoiesis. _Blood_ 113, 807–815 (2009). CAS PubMed PubMed Central Google Scholar * Adolfsson, J. et al. Identification of Flt3+ lympho-myeloid stem cells lacking
erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. _Cell_ 121, 295–306 (2005). CAS PubMed Google Scholar * Heinzel, K., Benz, C., Martins, V. C.,
Haidl, I. D. & Bleul, C. C. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. _J. Immunol._ 178, 858–868 (2007).
CAS PubMed Google Scholar * Radtke, F., Macdonald, H. R. & Tacchini-Cottier, F. Regulation of innate and adaptive immunity by Notch. _Nature Rev. Immunol._ 13, 427–437 (2013). CAS
Google Scholar * Rothenberg, E. V. T cell lineage commitment: identity and renunciation. _J. Immunol._ 186, 6649–6655 (2011). CAS PubMed PubMed Central Google Scholar * Masuda, K. et
al. T cell lineage determination precedes the initiation of TCRβ rearrangement. _J. Immunol._ 179, 3699–3706 (2007). CAS PubMed Google Scholar * Yui, M. A., Feng, N. & Rothenberg, E.
V. Fine-scale staging of T cell lineage commitment in adult mouse thymus. _J. Immunol._ 185, 284–293 (2010). REFERENCES 27 AND 28 REVEAL THE TIMING OF T CELL LINEAGE COMMITMENT, SHOWING ITS
CLEAR SEPARATION FROM TCR-DEPENDENT EVENTS AND DEFINING ITS BASIS IN TERMS OF CHANGES IN REGULATORY GENE EXPRESSION. CAS PubMed PubMed Central Google Scholar * Taghon, T., Yui, M. A.
& Rothenberg, E. V. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. _Nature Immunol._ 8, 845–855 (2007). CAS Google Scholar *
Wong, S. H. et al. Transcription factor RORα is critical for nuocyte development. _Nature Immunol._ 13, 229–236 (2012). CAS Google Scholar * Zhang, J. A., Mortazavi, A., Williams, B. A.,
Wold, B. J. & Rothenberg, E. V. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. _Cell_ 149, 467–482 (2012). THIS PAPER
PRESENTS THE FIRST GENOME-WIDE ANALYSIS OF THE EPIGENETIC CHANGES AND TRANSCRIPTIONAL DYNAMICS OF EARLY T CELL DEVELOPMENT, USING CHIP–SEQ AND RNA SEQUENCING ASSAYS ACROSS A SUCCESSION OF
STAGES SPANNING T CELL LINEAGE COMMITMENT. Article CAS PubMed PubMed Central Google Scholar * Mingueneau, M. et al. The transcriptional landscape of αβ T cell differentiation. _Nature
Immunol._ 14, 619–632 (2013). THIS PAPER USES MICROARRAY ANALYSES TO SHOW THE TRANSCRIPTOMIC CHANGES THAT OCCUR ACROSS AN EXTENDED SPAN OF T CELL DEVELOPMENT. RESULTS WERE GENERATED BY THE
IMMUNOLOGICAL GENOME PROJECT, WHICH IS AN INVALUABLE PUBLICLY ACCESSIBLE SOURCE OF GENE EXPRESSION DATA FOR ALL STAGES OF IMMUNE CELL DEVELOPMENT. CAS Google Scholar * Hoffman, E. S. et
al. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development _in vivo_. _Genes Dev._ 10, 948–962 (1996). CAS PubMed
Google Scholar * Taghon, T., Yui, M. A., Pant, R., Diamond, R. A. & Rothenberg, E. V. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the
adult mouse thymus. _Immunity_ 24, 53–64 (2006). CAS PubMed Google Scholar * Teague, T. K. et al. CD28 expression redefines thymocyte development during the pre-T to DP transition. _Int.
Immunol._ 22, 387–397 (2010). CAS PubMed Google Scholar * Taghon, T. et al. Notch signaling is required for proliferation but not for differentiation at a well-defined β-selection
checkpoint during human T-cell development. _Blood_ 113, 3254–3263 (2009). CAS PubMed Google Scholar * Kreslavsky, T. et al. β-Selection-induced proliferation is required for αβ T cell
differentiation. _Immunity_ 37, 840–853 (2012). CAS PubMed PubMed Central Google Scholar * Ciofani, M., Knowles, G. C., Wiest, D. L., von Boehmer, H. & Zúñiga-Pflücker, J. C.
Stage-specific and differential Notch dependency at the αβ and γδ T lineage bifurcation. _Immunity_ 25, 105–116 (2006). CAS PubMed Google Scholar * Maillard, I. et al. The requirement for
Notch signaling at the β-selection checkpoint _in vivo_ is absolute and independent of the pre-T cell receptor. _J. Exp. Med._ 203, 2239–2245 (2006). CAS PubMed PubMed Central Google
Scholar * Garbe, A. I. & von Boehmer, H. TCR and Notch synergize in αβ versus γδ lineage choice. _Trends Immunol._ 28, 124–131 (2007). CAS PubMed Google Scholar * Kueh, H. Y. &
Rothenberg, E. V. Regulatory gene network circuits underlying T cell development from multipotent progenitors. _Wiley Interdiscip. Rev. Syst. Biol. Med._ 4, 79–102 (2012). CAS PubMed
Google Scholar * Naito, T., Tanaka, H., Naoe, Y. & Taniuchi, I. Transcriptional control of T-cell development. _Int. Immunol._ 23, 661–668 (2011). CAS PubMed Google Scholar * Mercer,
E. M., Lin, Y. C. & Murre, C. Factors and networks that underpin early hematopoiesis. _Semin. Immunol._ 23, 317–325 (2011). PubMed PubMed Central Google Scholar * Rothenberg, E. V.
Transcriptional drivers of the T-cell lineage program. _Curr. Opin. Immunol._ 24, 132–138 (2012). CAS PubMed PubMed Central Google Scholar * De Pooter, R. F. & Kee, B. L. E proteins
and the regulation of early lymphocyte development. _Immunol. Rev._ 238, 93–109 (2010). CAS PubMed PubMed Central Google Scholar * Braunstein, M. & Anderson, M. K. HEB in the
spotlight: Transcriptional regulation of T-cell specification, commitment, and developmental plasticity. _Clin. Dev. Immunol._ 2012, 678–705 (2012). Google Scholar * Rothenberg, E. V.,
Zhang, J. & Li, L. Multilayered specification of the T-cell lineage fate. _Immunol. Rev._ 238, 150–168 (2010). CAS PubMed PubMed Central Google Scholar * David-Fung, E. S. et al.
Transcription factor expression dynamics of early T-lymphocyte specification and commitment. _Dev. Biol._ 325, 444–467 (2009). CAS PubMed Google Scholar * Tabrizifard, S. et al. Analysis
of transcription factor expression during discrete stages of postnatal thymocyte differentiation. _J. Immunol._ 173, 1094–1102 (2004). CAS PubMed Google Scholar * Kawazu, M. et al.
Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. _J. Immunol._ 179, 5335–5345 (2007). CAS PubMed Google
Scholar * Manesso, E., Chickarmane, V., Kueh, H. Y., Rothenberg, E. V. & Peterson, C. Computational modelling of T-cell formation kinetics: output regulated by initial
proliferation-linked deferral of developmental competence. _J. R. Soc. Interface_ 10, 20120774 (2013). PubMed PubMed Central Google Scholar * Gwin, K. A., Shapiro, M. B., Dolence, J. J.,
Huang, Z. L. & Medina, K. L. Hoxa9 and Flt3 signaling synergistically regulate an early checkpoint in lymphopoiesis. _J. Immunol._ 191, 745–754 (2013). CAS PubMed PubMed Central
Google Scholar * Huang, Y. et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. _Blood_ 119, 388–398 (2012). CAS PubMed PubMed Central Google Scholar
* Riddell, J. et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. _Cell_ 157, 549–564 (2014). CAS PubMed PubMed Central Google
Scholar * Yu, Y. et al. Bcl11a is essential for lymphoid development and negatively regulates p53. _J. Exp. Med._ 209, 2467–2483 (2012). CAS PubMed PubMed Central Google Scholar *
Capron, C. et al. The _SCL_ relative _LYL-1_ is required for fetal and adult hematopoietic stem cell function and B-cell differentiation. _Blood_ 107, 4678–4686 (2006). CAS PubMed Google
Scholar * Souroullas, G. P., Salmon, J. M., Sablitzky, F., Curtis, D. J. & Goodell, M. A. Adult hematopoietic stem and progenitor cells require either _Lyl1_ or _Scl_ for survival.
_Cell Stem Cell_ 4, 180–186 (2009). CAS PubMed PubMed Central Google Scholar * Zohren, F. et al. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of
early T lineage progenitors. _Nature Immunol._ 13, 761–769 (2012). CAS Google Scholar * McCormack, M. P. et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL.
_Blood_ 122, 2093–2103 (2013). CAS PubMed Google Scholar * Lécuyer, E. et al. The SCL complex regulates _c-kit_ expression in hematopoietic cells through functional interaction with Sp1.
_Blood_ 100, 2430–2440 (2002). PubMed Google Scholar * Phelan, J. D. et al. Growth factor independent-1 maintains Notch1-dependent transcriptional programming of lymphoid precursors. _PLoS
Genet._ 9, e1003713 (2013). CAS PubMed PubMed Central Google Scholar * Thoms, J. A. I. et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic
cells by a stem cell enhancer. _Blood_ 117, 7079–7089 (2011). CAS PubMed Google Scholar * McCormack, M. P. et al. The _Lmo2_ oncogene initiates leukemia in mice by inducing thymocyte
self-renewal. _Science_ 327, 879–883 (2010). IN THIS PAPER, THE AUTHORS USE FATE MAPPING TO DEMONSTRATE THE ROLE OF _LMO2_ IN INDUCING SELF-RENEWAL IN EARLY T CELLS, WHICH CAN LEAD TO
LEUKAEMIA INITIATION. THIS ROLE OF _LMO2_ IS A PROTOTYPE FOR LINKING NATURAL THYMOCYTE PROLIFERATIVE EXPANSION WITH ONCOGENESIS. CAS PubMed Google Scholar * Carotta, S., Wu, L. &
Nutt, S. L. Surprising new roles for PU.1 in the adaptive immune response. _Immunol. Rev._ 238, 63–75 (2010). CAS PubMed Google Scholar * Dakic, A. et al. PU.1 regulates the commitment of
adult hematopoietic progenitors and restricts granulopoiesis. _J. Exp. Med._ 201, 1487–1502 (2005). CAS PubMed PubMed Central Google Scholar * Del Real, M. M. & Rothenberg, E. V.
Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. _Development_ 140, 1207–1219 (2013). CAS PubMed PubMed Central Google Scholar * Heinz, S. et al.
Effect of natural genetic variation on enhancer selection and function. _Nature_ 503, 487–492 (2013). CAS PubMed PubMed Central Google Scholar * Heinz, S. et al. Simple combinations of
lineage-determining transcription factors prime _cis_-regulatory elements required for macrophage and B cell identities. _Mol. Cell_ 38, 576–589 (2010). CAS PubMed PubMed Central Google
Scholar * Ostuni, R. & Natoli, G. Lineages, cell types and functional states: a genomic view. _Curr. Opin. Cell Biol._ 25, 759–764 (2013). CAS PubMed Google Scholar * Ghisletti, S.
et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. _Immunity_ 32, 317–328 (2010). CAS PubMed Google Scholar *
Visan, I. et al. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. _Nature Immunol._ 7, 634–643 (2006). CAS Google Scholar *
Georgescu, C. et al. A gene regulatory network armature for T lymphocyte specification. _Proc. Natl Acad. Sci. USA_ 105, 20100–20105 (2008). CAS PubMed Google Scholar * Weng, A. P. et al.
_c-Myc_ is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. _Genes Dev._ 20, 2096–2109 (2006). CAS PubMed PubMed Central Google Scholar * Wang, D.
et al. The basic helix–loop–helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. _J. Immunol._ 177, 109–119 (2006). CAS PubMed
Google Scholar * De Obaldia, M. E. et al. T cell development requires constraint of the myeloid regulator C/EBP-α by the Notch target and transcriptional repressor Hes1. _Nature Immunol._
14, 1277–1284 (2013). CAS Google Scholar * Wendorff, A. A. et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and
transformation. _Immunity_ 33, 671–684 (2010). REFERENCES 75 AND 76 DEMONSTRATE THE CRUCIAL IMPORTANCE OF _HES1_ AS A NOTCH TARGET GENE IN THE EARLIEST STAGES OF T CELL DEVELOPMENT. TOGETHER
THESE REFERENCES SHOW ITS DISCRETE ROLE IN COMMITMENT (REFERENCE 75) AND IN NORMAL POPULATION EXPANSION, AS WELL AS IN NOTCH-INDUCED T-ALL (REFERENCE 76). CAS PubMed Google Scholar *
Tomita, K. et al. The bHLH gene _Hes1_ is essential for expansion of early T cell precursors. _Genes Dev._ 13, 1203–1210 (1999). CAS PubMed PubMed Central Google Scholar * Tsuji, M.,
Shinkura, R., Kuroda, K., Yabe, D. & Honjo, T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J
signaling. _Proc. Natl Acad. Sci. USA_ 104, 1610–1615 (2007). CAS PubMed Google Scholar * Yun, T. J. & Bevan, M. J. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling:
multiple Notch1 signaling pathways involved in T cell development. _J. Immunol._ 170, 5834–5841 (2003). CAS PubMed Google Scholar * Hosoya, T. et al. GATA-3 is required for early T
lineage progenitor development. _J. Exp. Med._ 206, 2987–3000 (2009). CAS PubMed PubMed Central Google Scholar * Germar, K. et al. T-cell factor 1 is a gatekeeper for T-cell
specification in response to Notch signaling. _Proc. Natl Acad. Sci. USA_ 108, 20060–20065 (2011). CAS PubMed Google Scholar * Miyazaki, M. et al. The opposing roles of the transcription
factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. _Nature Immunol._ 12, 992–1001 (2011). CAS Google Scholar * Ikawa, T., Kawamoto, H., Goldrath, A.
W. & Murre, C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. _J. Exp. Med._ 203, 1329–1342 (2006). CAS PubMed PubMed Central Google
Scholar * Engel, I. & Murre, C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. _EMBO J._ 23, 202–211 (2004). CAS PubMed Google Scholar * Jones-Mason, M. E.
et al. E protein transcription factors are required for the development of CD4+ lineage T cells. _Immunity_ 36, 348–361 (2012). CAS PubMed PubMed Central Google Scholar * Jones, M. E.
& Zhuang, Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. _Immunity_ 27, 860–870 (2007). CAS PubMed
PubMed Central Google Scholar * Weber, B. N. et al. A critical role for TCF-1 in T-lineage specification and differentiation. _Nature_ 476, 63–68 (2011). REFERENCES 81 AND 87 DEMONSTRATE
THAT TCF1 IS A KEY MEDIATOR OF T CELL SPECIFICATION THAT FUNCTIONS DOWNSTREAM OF NOTCH SIGNALLING. REFERENCE 87 SHOWS THAT HIGH LEVELS OF TCF1 DRIVE THE EXPRESSION OF T CELL GENES INCLUDING
_GATA3_ AND _BCL11B_ EVEN IN THE ABSENCE OF NOTCH SIGNALS. CAS PubMed PubMed Central Google Scholar * Tiemessen, M. M. et al. The nuclear effector of Wnt-signaling, Tcf1, functions as a
T-cell-specific tumor suppressor for development of lymphomas. _PLoS Biol._ 10, e1001430 (2012). CAS PubMed PubMed Central Google Scholar * Yu, S. et al. The TCF-1 and LEF-1
transcription factors have cooperative and opposing roles in T cell development and malignancy. _Immunity_ 37, 813–826 (2012). REFERENCES 88 AND 89 SHOW THE COMPLEX INTRA-FAMILY
RELATIONSHIPS BETWEEN TCF1 AND LEF1 IN EARLY T CELL DEVELOPMENT, AND THE IMPORTANCE OF CERTAIN ISOFORMS OF TCF1 THAT FUNCTION AS TUMOUR SUPPRESSORS BY REPRESSING THE EXPRESSION OF _LEF1_.
CAS PubMed PubMed Central Google Scholar * Schilham, M. W. et al. Critical involvement of Tcf-1 in expansion of thymocytes. _J. Immunol._ 161, 3984–3991 (1998). CAS PubMed Google
Scholar * Staal, F. J. T. & Sen, J. M. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. _Eur. J. Immunol._ 38, 1788–1794 (2008). CAS
PubMed PubMed Central Google Scholar * Dose, M. et al. β-Catenin induces T-cell transformation by promoting genomic instability. _Proc. Natl Acad. Sci. USA_ 111, 391–396 (2014). CAS
PubMed Google Scholar * Giese, K., Kingsley, C., Kirshner, J. R. & Grosschedl, R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and
multiple protein-protein interactions. _Genes Dev._ 9, 995–1008 (1995). CAS PubMed Google Scholar * Ji, H. et al. Comprehensive methylome map of lineage commitment from haematopoietic
progenitors. _Nature_ 467, 338–342 (2010). CAS PubMed PubMed Central Google Scholar * Hosoya, T., Maillard, I. & Engel, J. D. From the cradle to the grave: activities of GATA-3
throughout T-cell development and differentiation. _Immunol. Rev._ 238, 110–125 (2010). CAS PubMed PubMed Central Google Scholar * Ho, I. C., Tai, T. S. & Pai, S. Y. GATA3 and the
T-cell lineage: essential functions before and after T-helper-2-cell differentiation. _Nature Rev. Immunol._ 9, 125–135 (2009). CAS Google Scholar * García-Ojeda, M. E. et al. GATA-3
promotes T cell specification by repressing B cell potential in pro-T cells. _Blood_ 121, 1749–1759 (2013). THIS PAPER IDENTIFIES GATA3 AS THE CRUCIAL INTRINSIC REGULATORY FACTOR IN THE
EARLIEST T CELL LINEAGE PRECURSOR CELLS THAT IS RESPONSIBLE FOR EXCLUDING ACCESS TO THE B CELL LINEAGE. PubMed Google Scholar * Tan, J. B., Visan, I., Yuan, J. S. & Guidos, C. J.
Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. _Nature Immunol._ 6, 671–679 (2005). CAS Google Scholar * Sambandam, A. et al. Notch signaling
controls the generation and differentiation of early T lineage progenitors. _Nature Immunol._ 6, 663–670 (2005). CAS Google Scholar * Pai, S. Y. et al. Critical roles for transcription
factor GATA-3 in thymocyte development. _Immunity_ 19, 863–875 (2003). CAS PubMed Google Scholar * Hosoya-Ohmura, S. et al. An NK and T cell enhancer lies 280 kilobase pairs 3′ to the
_Gata3_ structural gene. _Mol. Cell. Biol._ 31, 1894–1904 (2011). CAS PubMed PubMed Central Google Scholar * Weishaupt, H., Sigvardsson, M. & Attema, J. L. Epigenetic chromatin
states uniquely define the developmental plasticity of murine hematopoietic stem cells. _Blood_ 115, 247–256 (2010). CAS PubMed Google Scholar * Vigano, M. A. et al. An epigenetic profile
of early T-cell development from multipotent progenitors to committed T-cell descendants. _Eur. J. Immunol._ 44, 1181–1193 (2014). CAS PubMed Google Scholar * Xu, W. et al. E2A
transcription factors limit expression of _Gata3_ to facilitate T lymphocyte lineage commitment. _Blood_ 121, 1534–1542 (2013). THIS PAPER REVEALS THAT GATA3 ACTIVITY MUST BE KEPT UNDER
INHIBITORY RESTRAINT BY THE SAME BHLH E PROTEINS THAT ALSO COLLABORATE WITH IT TO DRIVE T CELL SPECIFICATION. THIS IS SHOWN TO BE ONE IMPORTANT WAY IN WHICH E2A PROMOTES SUCCESSFUL T CELL
COMMITMENT. CAS PubMed PubMed Central Google Scholar * Xu, W. & Kee, B. L. Growth factor independent 1B (_Gfi1b_) is an E2A target gene that modulates _Gata3_ in T-cell lymphomas.
_Blood_ 109, 4406–4414 (2007). CAS PubMed Google Scholar * Maneechotesuwan, K. et al. Regulation of TH2 cytokine genes by p38 MAPK-mediated phosphorylation of GATA-3. _J. Immunol._ 178,
2491–2498 (2007). CAS PubMed Google Scholar * Cook, K. D. & Miller, J. TCR-dependent translational control of GATA-3 enhances TH2 differentiation. _J. Immunol._ 185, 3209–3216 (2010).
CAS PubMed PubMed Central Google Scholar * Frelin, C. et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. _Nature Immunol._ 14, 1037–1044 (2013). CAS Google
Scholar * Wei, G. et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. _Immunity_ 35, 299–311 (2011). CAS PubMed PubMed Central
Google Scholar * Okamura, R. M. et al. Redundant regulation of T cell differentiation and TCRβ gene expression by the transcription factors LEF-1 and TCF-1. _Immunity_ 8, 11–20 (1998). CAS
PubMed Google Scholar * Wojciechowski, J., Lai, A., Kondo, M. & Zhuang, Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. _J. Immunol._ 178,
5717–5726 (2007). CAS PubMed PubMed Central Google Scholar * Zhong, Y., Jiang, L., Hiai, H., Toyokuni, S. & Yamada, Y. Overexpression of a transcription factor _LYL1_ induces T- and
B-cell lymphoma in mice. _Oncogene_ 26, 6937–6947 (2007). CAS PubMed Google Scholar * Welinder, E. et al. The transcription factors E2A and HEB act in concert to induce the expression of
FOXO1 in the common lymphoid progenitor. _Proc. Natl Acad. Sci. USA_ 108, 17402–17407 (2011). CAS PubMed Google Scholar * Schwartz, R., Engel, I., Fallahi-Sichani, M., Petrie, H. T. &
Murre, C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. _Proc. Natl Acad. Sci. USA_ 103, 9976–9981
(2006). CAS PubMed Google Scholar * Takeuchi, A. et al. E2A and HEB activate the pre-TCRα promoter during immature T cell development. _J. Immunol._ 167, 2157–2163 (2001). CAS PubMed
Google Scholar * Ikawa, T. et al. An essential developmental checkpoint for production of the T cell lineage. _Science_ 329, 93–96 (2010). CAS PubMed Google Scholar * Li, L., Leid, M.
& Rothenberg, E. V. An early T cell lineage commitment checkpoint dependent on the transcription factor _Bcl11b_. _Science_ 329, 89–93 (2010). CAS PubMed PubMed Central Google Scholar
* Li, P. et al. Reprogramming of T cells to natural killer-like cells upon _Bcl11b_ deletion. _Science_ 329, 85–89 (2010). REFERENCES 116–118 DEFINE THE ROLE OF _BCL11B_ IN T CELL LINEAGE
COMMITMENT. CAS PubMed PubMed Central Google Scholar * Cismasiu, V. B. et al. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter.
_Oncogene_ 24, 6753–6764 (2005). CAS PubMed Google Scholar * Li, L. et al. A far downstream enhancer for murine _Bcl11b_ controls its T-cell specific expression. _Blood_ 122, 902–911
(2013). CAS PubMed PubMed Central Google Scholar * Guo, Y., Maillard, I., Chakraborti, S., Rothenberg, E. V. & Speck, N. A. Core binding factors are necessary for natural killer cell
development, and cooperate with Notch signaling during T cell specification. _Blood_ 112, 480–492 (2008). CAS PubMed PubMed Central Google Scholar * Kastner, P. et al. Bcl11b represses
a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. _Eur. J. Immunol._ 40, 2143–2154 (2010). CAS PubMed PubMed Central Google Scholar * Oosterwegel, M. et al.
Cloning of murine TCF-1, a T cell-specific transcription factor interacting with functional motifs in the CD3-ɛ and T cell receptor α enhancers. _J. Exp. Med._ 173, 1133–1142 (1991). CAS
PubMed Google Scholar * Yashiro-Ohtani, Y. et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. _Genes Dev._ 23, 1665–1676 (2009). CAS PubMed PubMed Central
Google Scholar * Lauritsen, J. P. et al. Marked induction of the helix-loop-helix protein Id3 promotes the γδ T cell fate and renders their functional maturation Notch independent.
_Immunity_ 31, 565–575 (2009). CAS PubMed PubMed Central Google Scholar * Wakabayashi, Y. et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. _Nature
Immunol._ 4, 533–539 (2003). CAS Google Scholar * Inoue, J. et al. Expression of TCRαβ partly rescues developmental arrest and apoptosis of αβ T cells in _Bcl11b__−/−_ mice. _J. Immunol._
176, 5871–5879 (2006). CAS PubMed Google Scholar * Shibata, K. et al. IFN-γ-producing and IL-17-producing γδ T cells differentiate at distinct developmental stages in murine fetal thymus.
_J. Immunol._ 192, 2210–2218 (2014). CAS PubMed Google Scholar * Barndt, R. J., Dai, M. & Zhuang, Y. Functions of E2A–HEB heterodimers in T-cell development revealed by a dominant
negative mutation of HEB. _Mol. Cell. Biol._ 20, 6677–6685 (2000). CAS PubMed PubMed Central Google Scholar * Winandy, S., Wu, L., Wang, J. H. & Georgopoulos, K. Pre-T cell receptor
(TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. _J. Exp. Med._ 190, 1039–1048 (1999). CAS PubMed PubMed Central Google Scholar * Ciofani, M. &
Zúñiga-Pflücker, J. C. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. _Nature Immunol._ 6, 881–888 (2005). CAS Google Scholar *
Schjerven, H. et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. _Nature Immunol._ 14, 1073–1083 (2013). CAS Google Scholar * Zhang, J.
et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. _Nature Immunol._ 13, 86–94 (2012). CAS Google Scholar *
Chari, S. & Winandy, S. Ikaros regulates Notch target gene expression in developing thymocytes. _J. Immunol._ 181, 6265–6274 (2008). CAS PubMed PubMed Central Google Scholar *
Kleinmann, E., Geimer Le Lay, A. S., Sellars, M., Kastner, P. & Chan, S. Ikaros represses the transcriptional response to Notch signaling in T-cell development. _Mol. Cell. Biol._ 28,
7465–7475 (2008). CAS PubMed PubMed Central Google Scholar * Geimer Le Lay, A. S. et al. The tumor suppressor Ikaros shapes the repertoire of Notch target genes in T cells. _Sci.
Signal._ 7, ra28 (2014). PubMed Google Scholar * Tussiwand, R. et al. The preTCR-dependent DN3 to DP transition requires Notch signaling, is improved by CXCL12 signaling and is inhibited
by IL-7 signaling. _Eur. J. Immunol._ 41, 3371–3380 (2011). CAS PubMed Google Scholar * Janas, M. L. et al. Thymic development beyond β-selection requires phosphatidylinositol 3-kinase
activation by CXCR4. _J. Exp. Med._ 207, 247–261 (2010). CAS PubMed PubMed Central Google Scholar * Xi, H., Schwartz, R., Engel, I., Murre, C. & Kersh, G. J. Interplay between RORγt,
Egr3, and E proteins controls proliferation in response to pre-TCR signals. _Immunity_ 24, 813–826 (2006). CAS PubMed Google Scholar * Yosef, N. et al. Dynamic regulatory network
controlling TH17 cell differentiation. _Nature_ 496, 461–468 (2013). CAS PubMed PubMed Central Google Scholar * Ciofani, M. et al. A validated regulatory network for TH17 cell
specification. _Cell_ 151, 289–303 (2012). CAS PubMed PubMed Central Google Scholar * He, Y. W. et al. Down-regulation of the orphan nuclear receptor RORγt is essential for T lymphocyte
maturation. _J. Immunol._ 164, 5668–5674 (2000). CAS PubMed Google Scholar * Sun, Z. et al. Requirement for RORγ in thymocyte survival and lymphoid organ development. _Science_ 288,
2369–2373 (2000). CAS PubMed Google Scholar * Wang, R. et al. Transcription factor network regulating CD4+CD8+ thymocyte survival. _Crit. Rev. Immunol._ 31, 447–458 (2011). CAS PubMed
PubMed Central Google Scholar * Pongracz, J. E., Parnell, S. M., Jones, T., Anderson, G. & Jenkinson, E. J. Overexpression of ICAT highlights a role for catenin-mediated canonical Wnt
signalling in early T cell development. _Eur. J. Immunol._ 36, 2376–2383 (2006). CAS PubMed Google Scholar * Xu, M., Sharma, A., Wiest, D. L. & Sen, J. M. Pre-TCR-induced β-catenin
facilitates traversal through β-selection. _J. Immunol._ 182, 751–758 (2009). CAS PubMed PubMed Central Google Scholar * Xu, M., Sharma, A., Hossain, M. Z., Wiest, D. L. & Sen, J. M.
Sustained expression of pre-TCR induced β-catenin in post-β-selection thymocytes blocks T cell development. _J. Immunol._ 182, 759–765 (2009). CAS PubMed PubMed Central Google Scholar *
Schroeder, J. H., Bell, L. S., Janas, M. L. & Turner, M. Pharmacological inhibition of glycogen synthase kinase 3 regulates T cell development _in vitro_. _PLoS ONE_ 8, e58501 (2013).
CAS PubMed PubMed Central Google Scholar * Aifantis, I., Raetz, E. & Buonamici, S. Molecular pathogenesis of T-cell leukaemia and lymphoma. _Nature Rev. Immunol._ 8, 380–390 (2008).
CAS Google Scholar * Uckun, F. M. et al. Clinical features and treatment outcome of childhood T-lineage acute lymphoblastic leukemia according to the apparent maturational stage of
T-lineage leukemic blasts: a Children's Cancer Group study. _J. Clin. Oncol._ 15, 2214–2221 (1997). CAS PubMed Google Scholar * Ferrando, A. A. et al. Gene expression signatures
define novel oncogenic pathways in T cell acute lymphoblastic leukemia. _Cancer Cell_ 1, 75–87 (2002). CAS PubMed Google Scholar * Van Vlierberghe, P. et al. Prognostic relevance of
integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. _Blood_ 122, 74–82 (2013). CAS PubMed PubMed Central Google Scholar * Smith, S. et al. _LIM domain only-2_
(_LMO2_) induces T-cell leukemia by two distinct pathways. _PLoS ONE_ 9, e85883 (2014). PubMed PubMed Central Google Scholar * Haydu, J. E. & Ferrando, A. A. Early T-cell precursor
acute lymphoblastic leukaemia. _Curr. Opin. Hematol._ 20, 369–373 (2013). CAS PubMed Google Scholar * Van de Walle, I. et al. An early decrease in Notch activation is required for human
TCR-αβ lineage differentiation at the expense of TCR-γδ T cells. _Blood_ 113, 2988–2998 (2009). CAS PubMed Google Scholar * Joachims, M. L., Chain, J. L., Hooker, S. W., Knott-Craig, C.
J. & Thompson, L. F. Human αβ and γδ thymocyte development: TCR gene rearrangements, intracellular TCR β expression, and γδ developmental potential — differences between men and mice.
_J. Immunol._ 176, 1543–1552 (2006). CAS PubMed PubMed Central Google Scholar * Blom, B. & Spits, H. Development of human lymphoid cells. _Annu. Rev. Immunol._ 24, 287–320 (2006).
CAS PubMed Google Scholar * Aster, J. C., Blacklow, S. C. & Pear, W. S. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. _J. Pathol._
223, 262–273 (2011). CAS PubMed Google Scholar * Tremblay, C. S. & Hoang, T. Early T cell differentiation lessons from T-cell acute lymphoblastic leukemia. _Prog. Mol. Biol. Transl.
Sci._ 92, 121–156 (2010). CAS PubMed Google Scholar * Winandy, S., Wu, P. & Georgopoulos, K. A dominant mutation in the _Ikaros_ gene leads to rapid development of leukemia and
lymphoma. _Cell_ 83, 289–299 (1995). CAS PubMed Google Scholar * Weng, A. P. et al. Activating mutations of _NOTCH1_ in human T cell acute lymphoblastic leukemia. _Science_ 306, 269–271
(2004). CAS PubMed Google Scholar * Laurenti, E. et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. _Nature
Immunol._ 14, 756–763 (2013). CAS Google Scholar * Ntziachristos, P. et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. _Nature Med._
18, 298–301 (2012). CAS PubMed Google Scholar * Neumann, M. et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of _DNMT3A_ mutations. _Blood_ 121, 4749–4752 (2013). CAS
PubMed Google Scholar * Cleveland, S. M. et al. _Lmo2_ induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. _Stem Cells_ 31, 882–894 (2013). CAS
PubMed PubMed Central Google Scholar * Treanor, L. M. et al. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent
potential. _J. Exp. Med._ 211, 701–713 (2014). CAS PubMed PubMed Central Google Scholar * Yui, M. A. et al. Loss of T cell progenitor checkpoint control underlies leukemia initiation in
_Rag1_-deficient nonobese diabetic mice. _J. Immunol._ 190, 3276–3288 (2013). CAS PubMed PubMed Central Google Scholar * Yui, M. A. & Rothenberg, E. V. Deranged early T cell
development in immunodeficient strains of nonobese diabetic mice. _J. Immunol._ 173, 5381–5391 (2004). CAS PubMed Google Scholar * Litman, G. W., Anderson, M. K. & Rast, J. P.
Evolution of antigen binding receptors. _Annu. Rev. Immunol._ 17, 109–147 (1999). CAS PubMed Google Scholar * Vantourout, P. & Hayday, A. Six-of-the-best: unique contributions of γδ T
cells to immunology. _Nature Rev. Immunol._ 13, 88–100 (2013). CAS Google Scholar * Kang, J., Volkmann, A. & Raulet, D. H. Evidence that γδ versus αβ T cell fate determination is
initiated independently of T cell receptor signaling. _J. Exp. Med._ 193, 689–698 (2001). CAS PubMed PubMed Central Google Scholar * Melichar, H. J. et al. Regulation of γδ versus αβ T
lymphocyte differentiation by the transcription factor SOX13. _Science_ 315, 230–233 (2007). CAS PubMed Google Scholar * Feng, N., Vegh, P., Rothenberg, E. V. & Yui, M. A. Lineage
divergence at the first TCR-dependent checkpoint: preferential γδ and impaired αβ T cell development in nonobese diabetic mice. _J. Immunol._ 186, 826–837 (2011). CAS PubMed Google Scholar
* Van de Walle, I. et al. Specific Notch receptor–ligand interactions control human TCR-αβ/γδ development by inducing differential Notch signal strength. _J. Exp. Med._ 210, 683–697
(2013). CAS PubMed PubMed Central Google Scholar * Haks, M. C. et al. Attenuation of γδTCR signaling efficiently diverts thymocytes to the αβ lineage. _Immunity_ 22, 595–606 (2005). CAS
PubMed Google Scholar * Hayes, S. M., Li, L. & Love, P. E. TCR signal strength influences αβ/γδ lineage fate. _Immunity_ 22, 583–593 (2005). CAS PubMed Google Scholar * Narayan,
K. et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. _Nature Immunol._ 13, 511–518 (2012). CAS Google Scholar * Verykokakis, M. et al.
Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent “innate” γδ T cells. _PLoS ONE_ 5, e9303 (2010). PubMed PubMed Central Google Scholar *
Kreslavsky, T. et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. _Proc. Natl Acad. Sci. USA_ 106,
12453–12458 (2009). CAS PubMed Google Scholar * Kreslavsky, T. Gleimer, M. & von Boehmer, H. αβ versus γδ lineage choice at the first TCR-controlled checkpoint. _Curr. Opin. Immunol._
22, 185–192 (2010). CAS PubMed PubMed Central Google Scholar * Prinz, I., Silva-Santos, B. & Pennington, D. J. Functional development of γδ T cells. _Eur. J. Immunol._ 43, 1988–1994
(2013). CAS PubMed Google Scholar * Schlenner, S. M. & Rodewald, H. R. Early T cell development and the pitfalls of potential. _Trends Immunol._ 31, 303–310 (2010). CAS PubMed
Google Scholar * Richie Ehrlich, L. I., Serwold, T. & Weissman, I. L. _In vitro_ assays misrepresent _in vivo_ lineage potentials of murine lymphoid progenitors. _Blood_ 117, 2618–2624
(2011). PubMed PubMed Central Google Scholar * Benz, C. & Bleul, C. C. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. _J. Exp.
Med._ 202, 21–31 (2005). CAS PubMed PubMed Central Google Scholar * Laiosa, C. V., Stadtfeld, M., Xie, H., de Andres-Aguayo, L. & Graf, T. Reprogramming of committed T cell
progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. _Immunity_ 25, 731–744 (2006). CAS PubMed Google Scholar * Wölfler, A. et al. Lineage-instructive
function of C/EBPα in multipotent hematopoietic cells and early thymic progenitors. _Blood_ 116, 4116–4125 (2010). PubMed Google Scholar * Franco, C. B. et al. Notch/Delta signaling
constrains reengineering of pro-T cells by PU.1. _Proc. Natl Acad. Sci. USA_ 103, 11993–11998 (2006). CAS PubMed Google Scholar * Huang, G. et al. PU.1 is a major downstream target of
AML1 (RUNX1) in adult mouse hematopoiesis. _Nature Genet._ 40, 51–60 (2008). CAS PubMed Google Scholar * Zarnegar, M. A., Chen, J. & Rothenberg, E. V. Cell-type-specific activation
and repression of PU.1 by a complex of discrete, functionally specialized _cis_-regulatory elements. _Mol. Cell. Biol._ 30, 4922–4939 (2010). CAS PubMed PubMed Central Google Scholar *
Rosenbauer, F. et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. _Nature Genet._ 38, 27–37 (2006). CAS PubMed Google
Scholar * Hoyler, T. et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. _Immunity_ 37, 634–648 (2012). CAS PubMed PubMed Central
Google Scholar * Yang, Q. et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. _Immunity_ 38, 694–704 (2013). CAS PubMed PubMed Central Google Scholar *
Gentek, R. et al. Modulation of signal strength switches Notch from an inducer of T cells to an inducer of ILC2. _Front. Immunol._ 4, 334 (2013). PubMed PubMed Central Google Scholar *
Heng, T. S. P., Painter, M. W. & The Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. _Nature Immunol._ 9,
1091–1094 (2008). CAS Google Scholar Download references ACKNOWLEDGEMENTS The authors apologize to colleagues whose work helped to inspire this Review but could not be cited due to space
constraints. The authors thank present and former members of their group whose published and unpublished data, as well as helpful discussion, shaped the ideas presented here. The authors
gratefully acknowledge support from the US National Institutes of Health (NIH grant AI064590 to M.A.Y., and AI083514, AI095943 and HD076915 to E.V.R.) and the Albert Billings Ruddock
Professorship (to E.V.R). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Division of Biology 156–29, California Institute of Technology, Pasadena, 91125, California, USA Mary A. Yui &
Ellen V. Rothenberg Authors * Mary A. Yui View author publications You can also search for this author inPubMed Google Scholar * Ellen V. Rothenberg View author publications You can also
search for this author inPubMed Google Scholar ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. RELATED LINKS FURTHER INFORMATION ImmGen
POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR TABLE 1 POWERPOINT SLIDE FOR TABLE 2 SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION S1 (TABLE) Critical T cell specification transcription factor genes in murine early T cells and their progenitors (PDF 638 kb) SUPPLEMENTARY INFORMATION S2 (TABLE)
Critical phase 1-specific transcription factor genes in murine early T cells and their progenitors (PDF 633 kb) GLOSSARY * Commitment The stage at which cells give up their intrinsic
capability to produce more than one kind of descendant. This concept depends on recognizing that precursor cells begin with the intrinsic potential to give rise to various different types of
descendant, but the actual fate choices that such precursor cells adopt will be different depending on the signals that they receive from the environment. Commitment is the developmental
transition within a given pathway during which the chosen cell fate becomes intrinsically irreversible, independent of the environment. * Gene regulatory network A system of relationships
between a set of regulatory genes and the transcription factors that they encode, which is defined such that the interactions between them explain the stability or change in the
developmental properties of a cell type that expresses those genes. * Notch signalling A signalling system comprised of highly conserved transmembrane receptors that regulate cell fate
choice in the development of many cell lineages and are thus crucial for the regulation of embryonic differentiation and development. Unusually among signalling systems, the cytoplasmic
domain of each Notch transmembrane protein can itself become a transcriptional co-activator in the nucleus, as it can be proteolytically cleaved from the transmembrane region when Notch
interacts with its ligands of the Delta or Jagged family. * T cell acute lymphoblastic leukaemia (T-ALL). Leukaemia with an immature T cell phenotype. * Common lymphoid precursors (CLPs).
These are a type of progenitor cell that seems to be committed to lymphoid fates (as measured by _in vivo_ transfer) and that can give rise to all lymphoid cell types, including T cells, B
cells and natural killer cells. * Lymphoid-primed multipotent precursors (LMPPs). These are multilineage precursor cells that can generate myeloid and lymphoid descendants _in vivo_ and _in
vitro_ but that cannot generate erythroid or megakaryocytic cells. * Positive selection A step in the process of T cell differentiation in the thymus that selects CD4+CD8+ double-positive T
cells for survival and maturation, on the basis of the appropriate degree of interaction between their T cell receptor and the peptide—MHC complexes that are expressed on thymic epithelial
cells. Depending on the class of MHC molecule that is recognized, thymocytes are positively selected to a CD4+ or a CD8+ single-positive cell fate. * Pioneer factor A transcription factor
that has the ability to bind to its target site even when the site is initially located within nucleosome-packed chromatin, thus serving as a focal point for the recruitment of other
transcription factors. Pioneer factors are crucial for the multi-step process that is needed to activate some positive regulatory elements in the genome during development. * WNT A
signalling mediator named both for its mutant phenotype in _Drosophila melanogaster_ (Wingless) and for its role as a preferential retrovirus integration site in murine leukaemia
virus-induced leukaemias (Int-1). WNT signalling activates the T cell factor 1 (TCF1) and lymphoid enhancer-binding factor 1 (LEF1) family transcription factors through stabilizing their
co-activator, β-catenin, and mobilizing it from the cytoplasm to the nucleus. * Chromatin immunoprecipitation followed by sequencing (ChIP–seq). A genome-wide method of mapping the sites at
which a transcription factor binds to the DNA in a cell, that involves crosslinking proteins to chromatin, immunoprecipitating the chromatin with antibodies specific for the factor of
interest, comprehensively sequencing the DNA that is recovered in the immunoprecipitates and aligning the obtained sequences with the genome to identify the enriched regions. * Non-obese
diabetic mice (NOD mice). These mice spontaneously develop type 1 (insulin-dependent) diabetes mellitus as a result of autoreactive T cell-mediated destruction of pancreatic β-islet cells.
RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Yui, M., Rothenberg, E. Developmental gene networks: a triathlon on the course to T cell identity. _Nat
Rev Immunol_ 14, 529–545 (2014). https://doi.org/10.1038/nri3702 Download citation * Published: 25 July 2014 * Issue Date: August 2014 * DOI: https://doi.org/10.1038/nri3702 SHARE THIS
ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative
